Pseudomonas aeruginosa regulator PvrA binds simultaneously to multiple pseudo-palindromic sites for efficient transcription activation, a research team has found.
The study is led by Prof. Rui Bao and Prof. Zhaoming Su, Center of Infectious Diseases, Division of Infectious Diseases in State Key Laboratory of Biotherapy, West China Hospital, Sichuan University.
TetR family regulators (TFRs) are the largest group of DNA-binding transcription factors in bacteria and archaea, which play vital roles by regulating the expression of various genes governing diverse physiological pathways. Typically, TetRs negatively control the expression of target genes by competing with RNA polymerase.
TetR-type activators are relatively rare but are recently gaining an increasing interest because they play diverse roles in regulating the mechanisms underlying bacterial metabolism, stress, and virulence. Recently, a novel P. aeruginosa regulator PvrA was reported as a TetR-type activator that plays central roles in fatty acid metabolism.
DNA recognition
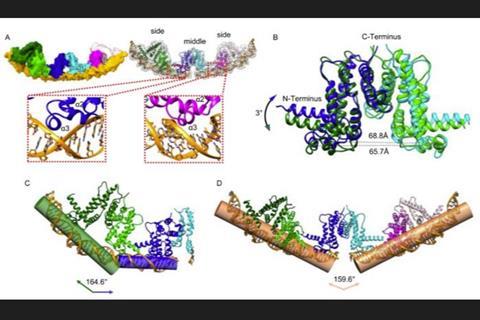
Zhu et al. reported that the DNA recognition properties of PvrA are more promiscuous than previously estimated. They showed that the presence of multiple pseudo-palindromic DNA sites binding within the target promoters is a significant mechanism underlying the complete activation effects of PvrA.
Results of cryo-electron microscopy (cryo-EM) analysis were used to recognize simultaneous DNA binding, and the results showed that the bound DNA fragments consisted of a distorted B-DNA double helix. Crystal structures of PvrA demonstrate the ligand-binding pocket of PvrA, and the giant cave permits the possibility of recognizing specific ligand governing different physiological pathways for global modulation.
Structural analyses reveal a unique hinge that secures the correct domain motion for promiscuous promoter recognition. They introduced mutations that disrupt the interactions in the regulatory hinge region, which exhibited differential effects on biofilm formation and pyocyanin biosynthesis, resulting in attenuated bacterial virulence.
These results indicate that PvrA via its unique regulatory mechanism enables P. aeruginosa to enhance its adaptability to environmental stress and virulence. This novel mechanism may improve the understanding of the structure-function relationship of TetR family and provide new insights into the mechanism governing the virulence of P. aeruginosa.





No comments yet