Persistent COPs such as tetrachloroethene and trichloroethene resist natural degradation, contaminating aquifers and soils for decades.
Microbial electrorespiration, which harnesses electrode-respiring bacteria to drive reductive dechlorination, is a promising solution. Yet, its application is hampered by aquifer heterogeneity, unpredictable microbial dynamics, and the difficulty of pinpointing the right operating conditions.
Current laboratory optimization often demands labor-intensive, unsustainable experiments. Meanwhile, machine learning has shown strong predictive power in fields such as water treatment and membrane design. Due to these challenges, there is an urgent need to develop data-driven strategies that can accelerate and scale up microbial dechlorination for contaminated environments.
Researchers from Harbin Institute of Technology and Northwestern Polytechnical University report a new machine learning framework that integrates experimental features with microbial biofilm data to optimize bioelectrodechlorination.
The study, published (DOI: 10.1016/j.ese.2025.100625) on September 27, 2025 in Environmental Science and Ecotechnology, demonstrates that inverse design can accurately predict dechlorination rates and identify the most effective conditions for breaking down chlorinated pollutants. By combining algorithms such as random forest and extreme gradient boosting with particle swarm optimization, the method reduces reliance on exhaustive laboratory testing while enhancing remediation efficiency.
Complex interplay
The team compiled a dataset of 357 entries from 68 peer-reviewed studies, covering experimental designs, cathodic biofilm profiles, and reaction rates. They tested multiple machine learning models, including random forest, multilayer perceptron, and XGBoost, to capture the complex interplay among environmental factors, electrochemical conditions, and microbial taxa.
Results showed that incorporating genus-level microbial data significantly improved predictions, with R² values above 0.87. Key drivers included temperature, cathode potential, electrode area, and the relative abundance of bacteria such as Clostridium, Desulfovibrio, Geobacter, and Dehalococcoides.
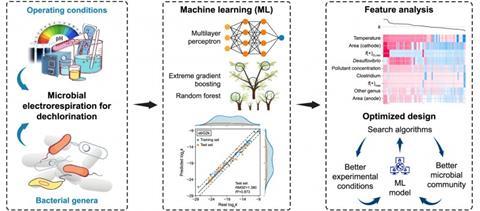
Using particle swarm optimization, the framework identified optimal potentials (−260 to −510 mV) and a temperature of ~23 °C for dechlorinating tetrachloroethene, trichloroethene, and 1,2-dichloroethane. Experimental tests confirmed that predictions matched observed reaction rates within 6% error.
Life cycle assessment further showed that adopting ML-guided optimization reduced global warming potential by nearly 15 kg CO₂-equivalent and lowered energy demand. This approach marks the first time microbial community data have been integrated into an inverse design model for bioelectrodechlorination, highlighting its potential to guide scalable, eco-friendly pollutant remediation.
Microbial ecology and machine learning
“Our study demonstrates that combining microbial ecology with machine learning can fundamentally change how we design bioremediation systems,” said corresponding author Prof. Aijie Wang.
“Instead of relying on lengthy trial-and-error experiments, we can now pinpoint effective operating conditions with high accuracy. This not only saves time and resources but also deepens our understanding of how microbial communities contribute to pollutant degradation. Such a framework provides a practical bridge between laboratory research and real-world environmental engineering.”
This data-driven framework paves the way for practical, scalable remediation of contaminated groundwater and soils. By cutting down experimental costs and improving predictive accuracy, the method accelerates the cleanup of sites polluted by industrial solvents, pesticides, and persistent chlorinated compounds.
Beyond COPs, the strategy can be adapted to other bioelectrochemical processes, including wastewater treatment, bioenergy production, and removal of emerging pollutants. As more datasets and genomic information become available, integrating functional genes with inverse design may further refine predictive models. Ultimately, this approach brings bioelectrochemical remediation closer to widespread, sustainable application in global environmental management.





No comments yet