Antimicrobial resistance (AMR) poses a significant threat to public health in the 21st century, with bacteria such as Campylobacter jejuni (C. jejuni) exhibiting multidrug resistance due to the presence of AMR genes. Understanding the evolutionary patterns and functional relationships of these genes is crucial for addressing this issue effectively.
Researchers in India have carried out a study, published in Gene Expression, which focused on tetracycline resistance genes tetO and tetM. By employing phylogenetic tree analysis, this research has provided valuable insights into the genetic landscape and variants associated with C. jejuni. The investigation highlighted the key hub genes such as rplE, rplV, rplG, and others, revealing their integral roles in AMR through GO keywords such as gene expression, cellular biosynthetic processes, and RNA binding.
Crucially, this study highlighted the significance of the rpl gene in driving the AMR phase of C. jejuni. These hub genes, exhibiting a high degree of clustering with their functional partners, have emerged as potential drug targets. The study’s findings raise hope that targeting these genes could pave the way for innovative treatments combating AMR in C. jejuni infections.
This comprehensive exploration of genetic and functional aspects offers valuable insights into the complex dynamics of AMR, providing a foundation for future therapeutic interventions and strategies in the ongoing battle against antibiotic resistance in C. jejuni.
Phylogenetic analysis
The authors conducted phylogenetic analysis to examine the evolution of AMR genes in C. jejuni. Additionally, they constructed and analyzed a gene interaction network comprising 39 functional relationships. Clustering analysis was employed to identify interconnected clusters associated with AMR processes. Functional enrichment analysis was performed to explore the involvement of cellular components, molecular functions, and biological processes.
The analysis revealed two interconnected clusters (C1 and C2) closely associated with AMR processes. Furthermore, genes encoding ribosomal proteins (rplE, rplV, rplG, rplK, rplA, rplJ, rpsE, rplB, rpsL, and rpmA) were identified as hub genes within the gene interaction network. These genes interact frequently with their functional counterparts, indicating their significance in AMR mechanisms. Enriched Kyoto Encyclopedia of Genes and Genomes pathway analysis highlighted the importance of the ribosome pathway in understanding antibiotic resistance mechanisms in C.
Potential drug targets
The investigation highlighted the key hub genes such as rplE, rplV, rplG, and others, revealing their integral roles in AMR through GO keywords such as gene expression, cellular biosynthetic processes, and RNA binding. Crucially, this study highlighted the significance of the rpl gene in driving the AMR phase of C. jejuni.
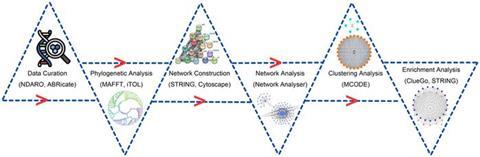
These hub genes, exhibiting a high degree of clustering with their functional partners, have emerged as potential drug targets. The study’s findings raise hope that targeting these genes could pave the way for innovative treatments combating AMR in C. jejuni infections.
This comprehensive exploration of genetic and functional aspects offers valuable insights into the complex dynamics of AMR, providing a foundation for future therapeutic interventions and strategies in the ongoing battle against antibiotic resistance in C. jejuni.





No comments yet