As a clinical academic, my research into mid-urethral mesh slings led me to wonder if the local microbiome played a role - and that meant developing a whole new skillset.
When I finished medical school, I knew I was interested in research and urogynaecology - the care of pelvic floor disorders like prolapse and incontinence - but I had virtually zero lab experience.
After medical school, I completed two years of foundation training in which all junior doctors rotate through 6 different specialities to gain basic competencies, and I completed three years of specialist training in Obstetrics and Gynaecology.
This specialist training pathway requires passing three postgraduate exams and takes seven years to complete, prior to becoming a consultant. Some of us follow slightly different pathways, to gain additional experience in research, education, leadership, and additional specialist skill sets.
Pause for research
Undertaking research for a higher degree, such as an MD or PhD, typically requires clinicians to pause clinical training. I worked clinically for half the week, in clinic, theatre and the delivery unit at St Mary’s Hospital, Manchester and in the McBain Microbiology lab for the other half of the week. The research pathway opens doors to becoming a clinical academic - someone who sits between clinical work and typically a university, to lead medical research.
The focus of my research was on mid-urethral mesh slings - a plastic, vaginally placed implant that supported the urethra to prevent leakage of urine secondary to an increase in abdominal pressure.

These implants have caused some women so much harm that their use has been paused in several countries and manufacturers have faced class-action lawsuits - these complications include mesh becoming exposed in the vagina, perforating into the bladder or urethra, and chronic pain.
Microbiome and mesh
My clinical supervisory team and I hypothesised that the local microbiome had an impact on mesh complications, and that biofilms were developing on mesh in vivo. We reached out to Prof McBain who we understood to be a biofilms expert, and built a new collaboration which we feel will continue to help improve women’s health.
Translating our hypothesis to surgeons and microbiologists gave me a new challenge - explaining what mesh complications were to microbiologists and the importance of the microbiome and biofilms to surgeons.

But I realised that delivering babies and fixing vaginal prolapses were not quite the same skillset as biofilm and microbiome research. I was fortunate to be welcomed into a very knowledgeable (and incredibly patient) microbiology lab headed by Professor Andrew McBain. Biggest shout-out to Tom Willmott who was quite saint-like whilst I learnt how to extract DNA, then repeated this with around 450 samples and undertook around 1,000 PCRs which were all checked by gel electrophoresis.
Muscle memory
The team joked that my surgical precision probably left me with a steady hand, making pipetting into tiny wells for electrophoresis easier; in reality, my hand usually shakes with an adrenaline surge after a complicated delivery.
I learnt that lab work requires huge organisation, focus and patience. The precision needs muscle memory and so it develops with experience - I am not sure how many thousand pipette tips I got through during my MD. Pipetting felt more like painting with a very fine brush than suturing skin - skin is much more forgiving than a misplaced pipette tip.
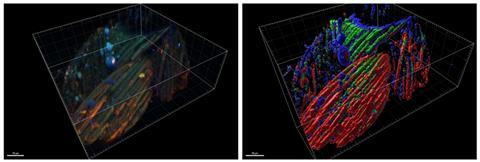
When DNA extractions got a bit tiring, I had a very welcome break with learning confocal laser scanning microscopy. We felt it was the best imaging modality for biofilms on explanted mesh, due to the 3D structure of the sample.
Pattern recognition
During medical school, I could never train my eyes to focus on a cell. Unfortunately, this just does not work when half your MD is imaging biofilms. After the first time I finally visualised a stained cell - thank you Cecile El Chami for telling me that was what I should be looking for - and my eyes seemed to learn the pattern quickly. I guess medical school trained us to become excellent at pattern recognition.
I was able to combine my clinical knowledge of the tissue samples with microbiology knowledge to understand that a Live/ Dead kit would not be suitable, and instead, we opted for a eubacterial FISH probe which had been used in the lab before.
I really felt that the translation of clinical to basic science, and vice versa, was starting to become my strength when considering our methodology. My feeling about microscopy and image rendering was that it really worked with the creative part of my brain - it didn’t really feel like work unless the sample floated away and refused to give me a nice image. I guess that is science.
Next learning curve
The next major learning curve was R studio - I have a newfound appreciation for those who code so remarkably well. I managed to pick up some basics and adapt the script which had been used for similar analyses.
It may be true that I would carry my laptop with an open lid to work over the weekend, plug it in in the doctor’s office of the gynaecology ward and return to it in between reviewing patients to make sure the code was still running… It was, it took 32 hours.
My average time for a straightforward caesarean section is about 30 minutes, so I could have delivered 64 babies in that time!
Biofilm findings
We found evidence of biofilms on all the explanted mesh we examined, as seen below, and that different complications were associated with change in microbiome at each relevant body site. Our findings have been exciting for the whole team, and we hope to continue this research to make mesh surgery safer for women or find new materials which do not result in the same complications.
My time being a pseudo-microbiology urogeynaecolgiosa has taught me that microbiology requires a great deal of patience, but when you see a little band at the right place on your gel, or generate a library after next-generation sequencing, it really pays off.
Becoming part of a team of clinicians and scientists has made me a better doctor. I can see the shift in my patient interactions when seeing consultations through the lens of a scientist - I am more curious and a better listener.
Dr Nazish Abbas MBBS, BSc (Hons) undertook an MD in Andrew McBain’s Microbiology lab, the University of Manchester, co-supervised by Dr Fiona Reid (Consultant Urogynaecologist, Warrell Unit, Saint Mary’s Hospital Manchester University NHS Foundation Trust) and Dr Maya Basu (Consultant Urogynaecologist, Warrell Unit, Saint Mary’s Hospital Manchester University NHS Foundation Trust).





No comments yet