A research team has proposed a new approach to reveal ecological niches (positions within ecosystems) and evolutionary relationships in nature through large-scale growth analysis of bacteria in strictly regulated laboratory settings.
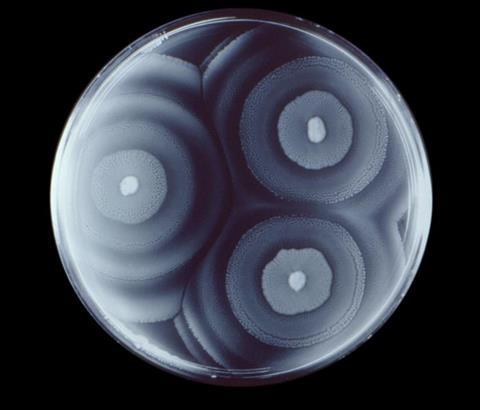
The researchers cultured six bacterial species in 195 different nutrient environments (media) and profiled their growth, generating 4,680 quantitative indices based on two parameters: growth rate and population size (maximum abundance). This experimental mapping provided a broad view of bacterial adaptation to diverse trophic environments.
READ MORE: Biologist examines growth patterns in bacteria to help develop more targeted, effective antibiotics
READ MORE: Scientists discover a materials maze that prevents bacterial infections
The results showed that while each species exhibited a distinct growth pattern, bacteria within the same phylogenetic lineage displayed similar trends. There were also consistent links with ecological traits such as habitat and distribution. Hierarchical clustering based on bacterial growth patterns is closely aligned with classifications derived from genomic information and ecological data from geographical distribution. This strong correlation demonstrates that growth profiles are conserved traits in both evolutionary and ecological contexts. Furthermore, the research highlighted two notable ways nutrients in culture media influence growth: the effects are either “universal” or “species-specific”.
The results suggest the feasibility of reproducing natural ecosystems and identifying laws of evolution and ecology through controlled experiments, using bacterial growth as an indicator. This study presents an alternative strategy for uncovering mechanisms of ecological and evolutionary adaptation in microorganisms. It also strengthens the ability to predict microbial adaptation and to refine classification methods with greater accuracy.
This work, published in Scientific Reports, was partially supported by JSPS KAKENHI (25K02259 to BWY) and JST SPRING (JPMJSP2124 to SZ).




No comments yet