Choice of alignment algorithms can make a huge difference to the outcome when you’re examining SARS-CoV-2 genome sequences, researchers will tell the Letters in Applied Microbiology ECS Research Symposium.
Dr Mohammed Abdallah Khodja from M’sila University in Algeria will be presenting his research, entitled ‘Pairwise sequence alignment analysis of Algerian SARS-COV2 Omicron’ at the event in Bristol, UK, this May.
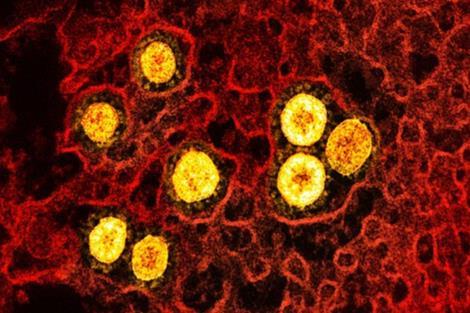
“In general, genomics sequence analysis for the purpose of genomic classification always poses problems,” he says.
“Firstly calculation problems, due to the long time; secondly, the cost problem, because efficient computation requires powerful computers, which can be expensive. Thirdly, algorithm improvement problems - to achieve accurate and optimal results, there’s a continuous need to improve algorithms used in genomic classification.”
Omicron in Algeria
To address these issues, the team conducted a case study focused on SARS-COV-2 Omicron in Algeria, using two methods - global and local sequence alignment.
The study on global and local pairwise alignment, focused on the effectiveness of the Needleman-Wunsch and Smith-Waterman algorithms for calculating matching scores. These algorithms are commonly used in bioinformatics for comparing biological sequences like DNA or protein sequences.
The research made use of real-life examples, specifically the SARS-CoV-2 genome sequences from different countries, which were obtained from the GISAID website. By applying these alignment algorithms, the researchers investigated the relationship between sequence structure, function, and evolution. They found that local alignment using the Spike region defined in the figure below, which focuses on finding regions of similarity within sequences, provided better grouping and classification of SARS-CoV-2 families.
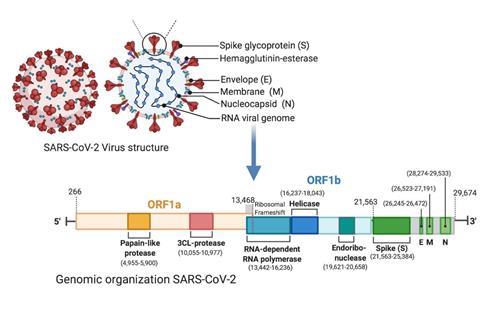
“We were surprised by the significant classification degradation, according to the Global vs Local alignment score,” Dr Khodja says.
“Indeed if we take our case as a model of treatment of genomic sequences, we can say that the genomic classification by Local sequence alignment is significantly better compared to Global alignment.”
Spike region
The importance of the spike region in the SARS-CoV-2 classification was highlighted by the results, and the study also discusses the challenges faced in computational biology, pointing out that even free platforms like Colab encounter difficulties when performing pairwise alignment on long sequences, such as the 30K nucleotide-long SARS-CoV-2 genome.
The researchers point to the need for researchers to ensure their samples, software, tools, and hardware are well-adapted for efficient genome analysis. They suggest that studies like this can be seen as initial steps toward using artificial intelligence to identify optimal sequence regions, which can be crucial for classifying microorganisms and conducting phylogenetic analyses. This is particularly important because genomic data are often very large, and standard computers may struggle to handle the necessary computations and analyses.
Real-world impacts
“Our work has two real-world impacts. Processing of small genomic sequences will save calculation time and should deliver accurate and optimal results,” Dr Khodja says.
“Incorrect classification may lead local or global health authorities, which as the World Health Organization, to make erroneous determinations of harm.This emphasizes the importance of using the most effective classification methods.
“In the next study, we hope to subject this case study to a larger-scale genomic data analysis by introducing artificial intelligence.”
This study was led by Dr Mohammed Abdallah Khodja, Assistant Professor in Bioinformatics at M’sila University with principal collaborator Dr Benazi Nabil who is responsible for the Pasteur Institut of Algeria. The work was carried out without any official budgetary funding.
The Letters in Applied Microbiology Early Career Scientist Research Symposium will be held at University of the West of England (UWE) in Bristol, UK, on 15 May 2024. To find out more, visit the event page.
Topics
- Applied Microbiology International
- Benazi Nabil
- Bioinformatics
- Community
- COVID-19
- Early Career Research
- M’sila University
- Middle East & Africa
- Mohammed Abdallah Khodja
- Needleman-Wunsch algorithm
- omicron
- One Health
- Pasteur Institut of Algeria
- Research News
- SARS-CoV-2
- Smith-Waterman algorithm
- Whole genome sequencing



No comments yet