We should start with a disclaimer that neither of us is a professional software developer or an artificial intelligence (AI) authority, but we have experience in programming, computational analysis of data, and are interested in early and successful adoption of AI for microbiology research. This is why we want to make a point that when it comes to AI, less is more.
The AI hype
AI is a loaded term, which is more often than not used interchangeably with machine learning (ML). Without going into semantic details and at the risk of invoking the ire of linguistic purists, we will use AI throughout the text as a broader definition. To a first approximation, AI can be divided into predictive AI, which processes input data and produces a prediction or classification based on the underlying model, and generative AI, which takes instructions and generates novel content as a result. Both types of AI can be extremely useful in microbiology research – if and when used correctly.
The public release of ChatGPT on 30 Nov 2022 made “AI” a truly household word. It can be argued that for biologists, this moment happened earlier, with the release of AlphaFold, which could predict protein folds with the accuracy rivalling that of X-ray crystallography. Artificial intelligence has swept into life sciences with a wave of promises. From decoding genomes to predicting protein structures and automating tedious benchwork, AI is heralded as the new engine of discovery. Convolutional neural networks can now count colonies or measure zones of inhibition faster than any PhD student with a marker pen. Reviews and data summaries can be produced almost instantaneously. We are currently living through an AI boom period. But as the AI fatigue slowly starts settling in, should microbiologists be racing to embed AI into every pipeline (as is happening in almost every other area of human endeavour)?
When it comes to the major issues with AI in microbiology, they can be broadly boiled down to three critical factors. 1) the Algorithm, 2) the Data, and 3) the User. As the majority of microbiologists are unlikely to develop their own AI solutions from scratch, here we are going to focus on the latter two.
The data: garbage in, garbage out
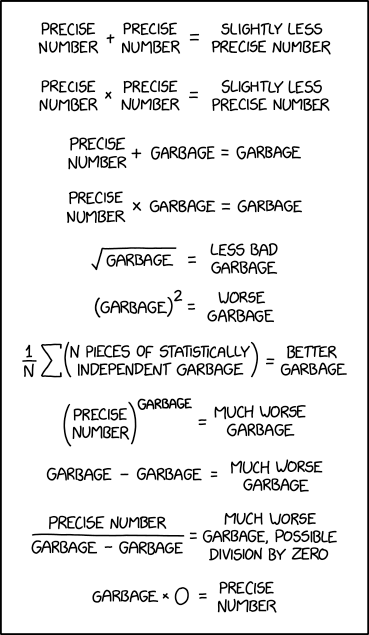
“More data beats clever algorithms, but better data beats more data”. This phrase, often attributed to Peter Norvig, one of the leading AI experts and former Director of Research at Google, describes the best approach to developing and using AI for microbiology research applications. Biological data is inherently complex and messy – more of it can help, but better preparation and careful curation help more.
AlphaFold exemplifies this approach. AlphaFold’s success came from training on a massive, expertly curated dataset: the Protein Data Bank - built from 50 years of painstaking structural biology. When similar models are applied to problems without that kind of data richness, they often stumble. Data limitations are acute – many microbiology datasets are “small‐n, large‐p” (few samples with very high-dimensional features), complicating model training and risking overfitting. For example, a 2024 study published in BMC Genomics reported that ML models trained to predict AMR in Escherichia coli utilising only whole genome sequencing datasets from England, demonstrated variable performance when tested on African isolates, suggesting that universal application of AI models and generalisation of findings to different settings should be carefully evaluated.
Machine learning is very good at finding correlations between data features, even in situations when such correlations have no biological meaning. Even when the amount of data used for training is sufficient, the resulting model would be prone to producing unreliable or outright erroneous predictions if the data is inaccurate, incomplete, biased, or contains artefacts. A high-profile example of this is a 2020 Nature paper in which highly accurate ML models (>95% accuracy) were developed to identify microbial DNA signatures discriminating cancer types. Unfortunately, despite the best efforts of the study authors to remove contaminating sequencing reads and avoid common pitfalls, a 2023 re-analysis revealed two major problems: misclassification of human sequences as being of bacterial origin and data normalisation giving rise to artificial signatures associated with specific cancer types, even in cases when these microorganisms were absent in the original dataset. These flaws invalidated the study’s findings and ultimately led to its retraction last year, but not before it had been widely cited and used as an inspiration for a number of related projects (and a couple of start-ups).
Unfortunately, the problems with data and metadata are pervasive in public databases. Many bacterial RefSeq genomes are contaminated with sequences of other bacteria, with more than 1% of genomes being heavily (≥5%) contaminated, while up to 80% of sequences in several enzyme super-families are wrongly annotated, such as S-2-hydroxyacid oxidases (EC 1.1.3.15) in the BRENDA database. Only a small proportion of proteins with functional annotations have been experimentally validated. Although we tend to expect that sequence and structure similarity to known enzymes would allow us to reliably assign a function, it is not always the case. For example, in a recent study, a set of depolymerase enzymes identified from Klebsiella prophages was tested on a panel of 119 capsule types. Out of 14 enzymes tested, five were found to target a different serotype than that of their bacterial host, demonstrating that computational predictions still require experimental validation. Metadata is as important as the data itself; during the COVID-19 pandemic, many thousands of viral genome sequences were submitted without accompanying metadata, making such sequences virtually useless for any subsequent analyses.
The lack of metadata details and the presence of errors and mis-annotations result in their propagation through databases, and the use of AI for classification and functional annotation of data exacerbates the problem even further. The training of AI models on data generated or annotated by other AI can lead to AI degeneration, similarly to how the LLM-generated content (‘AI slop’) propagation on the internet threatens to lead to the collapse of future models trained on such data. In 2023, Kim et al.’s Nature Comms DeepECtransformer predicted functions for 450 E. coli unknown proteins; a follow-up bioRxiv audit argues that ≥280 predictions copied existing UniProt errors, plus dozens of biologically impossible functions (e.g. E. coli making mycothiol). Supervised models inherit all label mistakes; “novel” discoveries could be simply recycled training labels.
The user: any sufficiently advanced incompetence is indistinguishable from malice
Even when the training data is good and the model is excellent, poor understanding of AI by microbiologists due to the lack of technical expertise, time to learn, and/or interest can lead to disappointing results.
The first question that should be asked is, do we really need AI to answer this? Quite often, a simple linear regression model would be comparable in performance to more sophisticated random forest and neural network approaches, especially in situations when the signal is mostly linear, while being easier to implement, evaluate, interpret, and maintain. Another example is taxonomic classification – according to a recent evaluation, database-based classification methods (e.g. Kraken 2) have higher accuracy than ML approaches (e.g. IDTAXA) when reference databases are comprehensive. In contrast, ML phage host range predictors outperform traditional alignment-based and alignment-free approaches, but accurate predictions appear to be mostly limited to genus- and species-level, making them useful for viral ecology studies but less so for selection of phage cocktails for phage therapy applications, which often require strain-level accuracy.
The second question is how to formulate the problem or correctly prepare data for AI-based analysis. The lack of understanding of how to properly configure and/or use AI would result in suboptimal performance. An example of that is the output of Deep Research, which substantially depends on the prompt, context, and examples provided to the AI prior to search. Quite often, using more structured prompts, asking another LLM to help generate a prompt, or trying reverse prompting (making the AI ask the user questions) substantially helps to improve the quality and relevance of the resulting Deep Research report.
Good understanding of AI limitations, biases, and pitfalls is critical. We have already mentioned data-related issues, such as data leakage from pre-training, catastrophic error propagation from incorrect labels, and overfitting to the training data. Predictive AI models can be fragile (their performance could vary substantially depending on the dataset used for training and could not be generalised) and tend to have inductive bias toward majority patterns, so rare classes are underweighted. Similarly to that, generative models gravitate towards specific, optimal solutions, sacrificing diversity in their output. Generative AI can be sensitive to prompts and/or context (lacking robustness), with even minor differences in the input data/query potentially leading to substantially different output and degraded performance. Erroneously constructed responses (whether textual, visual, or of some other type) are a notable problem with generative AI. These AI hallucinations (also known as confabulations, or according to a recent Nature article, simply “bullshit”) are quite common and widespread, with perhaps one of the most well-known and persisting issues being false scientific references. One early system (Galactica by Meta) specifically designed to help scientists to “summarize academic papers, solve math problems, generate Wiki articles, write scientific code, annotate molecules and proteins, and more” was pulled within three days after its release in November 2022 as it began producing fake citations and entirely fictional, albeit convincing, studies.
Finally, accidental or intentional lack of AI oversight, supervision, and QC of the results. LLMs tend to sound right and authoritative, so users assume machine output is correct and skip verification. Perhaps counterintuitively, the more reliable the model, the more dangerous its rare erroneous predictions or statements become, as it lulls the users into a false sense of security, making it more difficult (or virtually impossible) to notice inconsistencies and mistakes, especially for non-experts. It is not difficult to imagine a scenario when a software intended to detect antimicrobial genes in bacterial genomes, specific pathogen signatures in environmental samples, or for antibiotic prescription and which was developed with the help of a code generation AI such as GitHub Copilot would work correctly almost all the time, but fail in some edge cases, with potentially deadly consequences. This happened before the AI era (the infamous Therac-25 incidents killed at least three people), so the problem could be even worse due to the increasing popularity of vibe (AI-assisted) coding.
Some readers might have heard about a 2024 paper (now retracted) that featured several AI-generated figures, including an image of a particularly well-endowed rat. But the real issue wasn’t the figure; it was that nobody thought to question it, including the paper authors, the peer reviewers, and the editor of the journal. At the end of the day, the AI is a tool – it still requires a human to produce as well as inspect and approve the results.
Just a couple of days before the publication of this article, another problematic source was reported – this time, a whole book on ML with multiple citations that did not exist or had substantial errors (a hallmark of AI-hallucinated text). This invites the question: how many more erroneous results, fabricated citations, and hallucinated statements are lurking out there, poisoning our research literature corpus while remaining undetected?
Can AI simulate scientific thought?
We would like to digress here, as a few words should be said about reasoning models. Using AI to make predictions and discoveries runs into the limitation of what knowledge is included in its training data and what other information it can access (or generate) and incorporate into the reasoning process. We are talking about the world foundation model – the latent, internal representation of reality within the AI databanks, allowing it to build reasoning chains and solve problems. Similar, but smaller in scale, are foundation models for specific applications, including in biology (e.g. EVO-2, capable of generating genome fragments of up to 1 Mbp in size, and ESM3, enabling generation of protein structure and sequence based on the user’s prompt).
When we are solving a problem in biology, we interact with the physical, material world, with its underlying physical properties, processes, and laws, and reason based on the information we have. Frontier reasoning world models enable what is arguably the most advanced application of AI in research – the generation of new ideas and hypotheses, and planning of experiments with the help of so-called AI lab assistants or Co-scientists – MultiAgentic AI systems.
It should be mentioned that the potential ability of AI to simulate thinking processes remains contentious. A recent preprint from Apple AI (“The Illusion of Thinking”) argues that the existing large reasoning models are currently incapable of true thinking, experiencing accuracy collapse for problems beyond a certain complexity.
What is interesting is that although the AI hallucinations are a fundamental problem of LLMs, they might enable the simulation of a thinking, creative process, which perhaps is not dissimilar to how humans think. Albert Einstein once observed that “there is no logical bridge between phenomena and their theoretical principles”, or, in the words of the famous philosopher of science Paul Feyerabend, “anything goes”. To pose a problem, formulate a hypothesis, or generate a new idea, we string together what information we possess and then use our intuition, creativity, and ideas from other fields for closure of gaps – essentially hallucinating a novel idea from existing building blocks. The problem here is to limit these hallucinations in AI to steps where no reliable information about the problem/topic exists, and make such connections at least plausible.
AlphaFold fails to reliably predict protein structures of completely novel folds, absent from its training data. Even if AI cannot truly generate new ideas and simply acts as a kaleidoscope for known facts, rejiggling them in multiple ways, this still can be an invaluable tool for scientists, allowing them to quickly identify existing, but unseen connections in the existing data. Truly, the problem of thinking, reasoning, and creativity in AI is a fascinating one, but unfortunately, outside the scope of this article.
Five principles for responsible AI use
AI is extremely useful for making sense of large, complex datasets, and it is here to stay, but we need to be mindful of its (and our) limitations if we are to use it in a productive manner. We suggest applying the following principles:

1. RTFM (Read The Friendly Manual). An hour in the library saves a week in the lab, so before embarking on using a new model/software, read the accompanying paper and/or the user’s manual.

2. “All models are wrong, but some are useful.” Benchmark and compare the AI algorithms to each other and with traditional approaches. Be aware of the problems common to each of them.
3. Data is king. Make sure that the data you are planning to use is fit for purpose.

4. Be F.A.I.R. Apply F.A.I.R. principles (Findable, Accessible, Interoperable, and Reproducible) in your research. Upload your data to Zenodo and share your model on GitHub. “Data available on request” is un-F.A.I.R.
5. Trust, but verify. Even the best predictions are useless if they don’t work (or exist) in real life.







No comments yet