The viral world seems to be on the march. Novel coronaviruses have repetitively intruded into the human population with life-threatening diseases, first with SARS in 2002/2003, then with MERS starting in 2012 and lately culminating with the COVID-19 pandemic beginning in 2019.
The expanding viral world
Influenza viruses started with a fulminant pandemic, the Spanish flu from 1918, after which various major influenza virus epidemics followed. The Lentivirus HIV-1 caused the still ongoing AIDS pandemic in the 1980s. One can add further viruses - the paramyxovirus Nipahvirus, the Flavivirus Zikavirus, the monkeypox virus - to the list of novel viral threats. New viruses with epidemic potential are likely to emerge as a consequence of environmental and climate changes.
Pandemic preparedness
Assuring pandemic preparedness is thus an urgent public health task. Pandemic preparedness has many aspects and foreseeing the next ambush from the viral world is a steep challenge. Even predicting how ongoing viral epidemics will develop is notoriously difficult. SARS-CoV-2 is now said to have entered an endemic stage after the pandemic state. However, it is unclear what this will mean for the future development of SARS-CoV-2 infections. Will new viral strains evolve that undermine infection- and vaccine-acquired immunity? Instead of predicting the future from current trends, it might be worthwhile to look into historical records of past epidemics. The author of this feature has previously attempted a follow-up of the Spanish flu virus during the 20th century, through historical and virological records to retrace the later waves of an influenza virus pandemic. Another historical analysis of medical reports on the Russian flu pandemic from 1889 by the author revealed some striking clinical similarities with COVID-19 and might provide a possible framework for future developments of the COVID-19 epidemic. Your writer is a retired industrial microbiologist, now guest scientist at the Catholic University of Leuven and editor of Microbial Biotechnology where he has published a series of editorials under the name of “Lilliputs” that explore the impact of microbial research on our societies.
Endemic infections
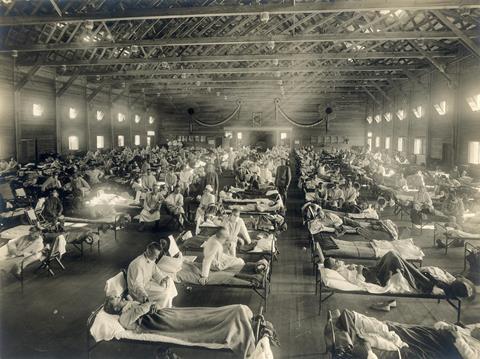
Endemic infections do not necessarily mean less dangerous infections. The orthopoxvirus Variola virus (VARV), the causative agent of smallpox, is a warning example for an endemic infection. In fact, VARV has no animal reservoir which was a crucial condition for its elimination by a WHO-directed eradication campaign through mass vaccination and case tracing followed by ring vaccination. In one of the major triumphs of modern medicine, VARV was declared extinct in 1980. Infected persons either recover by eliminating VARV and develop a sound immunity against reinfection or die from the disease. To persist in a human population VARV needs to maintain uninterrupted infection chains. Periodic epidemic phases depend on the addition of susceptible subjects through the birth of children devoid of immunity, influx of susceptible people by migration or spread to non-immune populations.
Impact and origin of smallpox
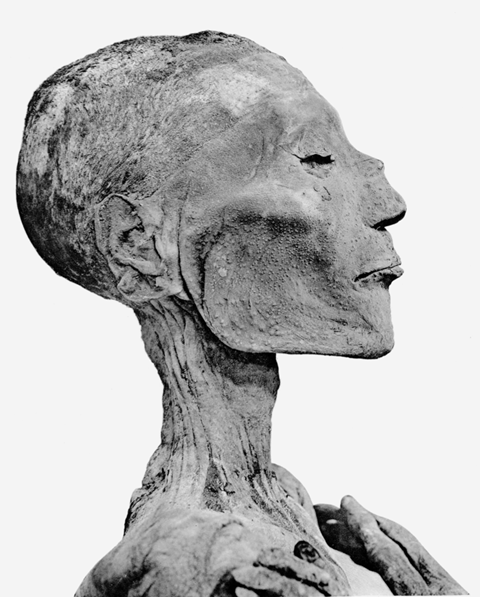
For all these aspects the history of smallpox is a fascinating illustration. It is most likely the greatest viral killer of human history. It was calculated that alone in the 20th century smallpox claimed more human lives than the Spanish flu pandemic, not to mention smallpox’s death toll in preceding centuries. Good epidemiological records exist for England: In the 18th century smallpox represented 10% of all deaths. In a particularly well documented study from Japan, smallpox was responsible for 2% of adult and 40% of childhood deaths between 1770 and 1850. Smallpox is an old disease. However, just how old it is has been a contentious issue until quite recently. Historians have pointed to the mummy of pharaoh Ramses V which showed pustules closely resembling smallpox. This would date smallpox virus in the human population back to 1157 BCE. This date, and presence in Egypt are not implausible since the closest virological relatives of VARV are found in camels and gerbils (a rodent), both living in arid areas. Camelpox is a severe veterinary disease, however gerbils only show asymptomatic infections. These observations suggest gerbils as a potential viral source and camels as intermediate host. Camels as pack animals were first used in the 12th century BCE in Southern Arabia and the Horn of Africa and apparently immediately used in copper mines of Ancient Egypt. Camels thus came into contact with one of the first human civilisations providing the necessary population density to maintain acute virus infection chains. The origin of smallpox in Egypt is therefore historically a plausible hypothesis. However, questions remain since smallpox was not mentioned in medical records from Ancient Egypt, nor by Hippocrates, the medical authority of Antiquity.
Spread of smallpox
The first medical reports clearly describing smallpox are from the 4th century CE (common era or AD) by a Chinese physician, from the 7th century in the Mediterranean area and the 10th century by a Persian physician. Smallpox might have travelled by trade from Egypt to the Levant and the Near East and by Buddhist missionaries from South to East Asia. Introduction into Europe was variously attributed to the invasion of the Huns, the expansion of Muslim warriors into Spain or crusaders returning from the Levant. Smallpox became catastrophic when it was introduced by Spanish conquistadores into the New World in the 16th century. Here viruses met a population that had no immunity and prior adaptation to these novel pathogens. Thirty years after the Spanish arrival about 30 million Indians had died from the newly introduced viruses. The historical records are, however, not precise enough to clearly differentiate between smallpox and measles infections. The slave trade from West Africa to America was a further motor for the spread of smallpox.
Variable virulence of smallpox
Europe received syphilis from America in exchange, which was then called “la grosse vérole” and distinguished from “la petite vérole” for smallpox (hence the English name). It is not clear whether petite vérole refers to smaller skin eruptions, the younger age of the victims or lesser virulence of smallpox compared to syphilis. Syphilis, when entering the Old World met an immunologically naïve population. However, the virulence of smallpox changed over time and smallpox became, with the stress of the Thirty Years War in Europe the major infectious disease in an exhausted population - outpacing plague, leprosy and syphilis.
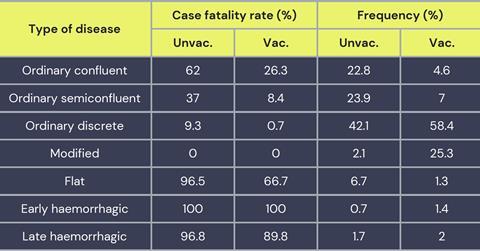
Throughout the 17th and 18th centuries smallpox claimed an enormous toll of human lives in Europe, which only began to decline with the early efforts of vaccination. The first vaccines were derived from the content of pustules of surviving victims, then with cowpox virus introduced by Jenner and later by vaccinia virus of unknown origin, culminating finally in the eradication of smallpox in 1980. However, VARV showed not only increased virulence development, but scientists also described milder forms of smallpox variously named as alastrim, variola minor or kaffir pox. Case fatality rates (CFR) for smallpox could thus vary from much more than 10% to much less than 1%, apparently associated with different viral strains. However, a large spectrum of clinical manifestations was also seen in defined outbreaks likely caused by a single viral strain. Disease severity ranged everywhere from the ‘ordinary’ and ‘modified’ categories, to the more severe ‘flat’ and ‘haemorrhagic’ smallpox. The stunning difference in case fatality rates (reaching more than 90% for flat and haemorrhagic smallpox) may reflect different host responses to the virus, rather than genetic differences between viral isolates. Pregnancy for example was defined as a prominent risk factor for elevated CFR with smallpox. The value of the historical records has been disputed but reports of the 10th century Persian physician Rhazes, or the 17th century British physician Sydenham provide clinical descriptions that fit closely with the symptomatology of smallpox as we know it, leaving no reasonable doubt that they dealt with authentic smallpox.
Ancient viral DNA analysis
Some ancient DNA analyses have provided excellent independent confirmations linking historical disease records with the genomes of known pathogens. The most spectacular case is perhaps the linking of the Black Death epidemic which depopulated 14th century Europe with the bacterium Yersinia pestis. One might expect a similar ancient genome insight for a DNA virus allowing to trace back smallpox into the historical past. However, the first sequencing efforts revealed a surprise. When two independent groups sequenced about 100 VARV isolates collected in the 30 years before smallpox eradication, two clades of VARV were identified. Phylogenetic tree analysis demonstrated that the ancestor of the contemporary VARV isolates existed only 220 years ago, and the two modern clades split just 90 years ago. What about the older historical records? Is smallpox as we know it from modern descriptions a relatively new disease? The conundrum of the age of smallpox virus was partially solved by modern mummies. First by an 18th century mummy buried in permafrost soil in Siberia and then by a child buried in 1650 in the crypt of a Lithuanian church. Lung histology from the permafrost mummy showed indications for hemorrhagic smallpox as cause of death. The mummies from both reports yielded VARV sequences that were ancestral to the two clades of modern VARV viruses. A few medical museum specimens linked these older examples to modern VARV isolates indicating a reliable molecular clock for this virus over the covered time period. Phylogenetic tree analysis of these viral sequences allowed us to trace back the ancestor of VARV into the 10th century CE.
The permafrost mummy yielded only very fragmented DNA complicating the genetic analysis. At the same time, the impossibility to amplify 2-kb long DNA fragments by PCR alleviates concerns that virulent VARV could be resurrected from thawing permafrost smallpox victims as a result of climate change, thereby kicking off an epidemic when encountering a now largely VARV-susceptible world population. Concerns for VARV as a bioterror agent remain, however. In another heroic effort ancient DNA was extracted from nearly 2000 human skeletons and teeth dated from 30,000-150 years ago and screened for VARV specific DNA. Twenty-six samples matched VARV sequences. Viral sequences could be enriched from 13 samples and 11 were from individuals who died between 603 CE and 1050 CE in northern Europe, western Russia, and the UK. Higher coverage VARV genome sequencing revealed the existence of a previously unknown, now-extinct VARV virus clade circulating in the Viking Age. The most recent common ancestor of these ancient VARVs and the modern VARVs was dated to the 4th century CE, approximately at the time when the term variola was first used by a bishop in Switzerland. With these genetic data the origin of VARV can at least be traced back to the earliest historical records describing smallpox by ancient physicians, although - so far - not back to Egyptian pharaohs.

The high coverage sequencing of the ancient VARV genomes allowed further interesting conclusions. Orthopoxvirus species with a narrow host range such as VARV have fewer genes than those with broad host ranges such as cowpox virus (CPXV) with 162 vs. 209 functional genes, respectively. The researchers identified a variable viral gene deletion pattern in the evolution of human VARV isolates that might - at least partly - underlie the changing virulence reported for smallpox in historical reports across time. For example, in the Viking Age only 2% of the skeletons showed evidence of smallpox infection while in 18th century Britain, 10-20% of deaths were caused by smallpox. During the Viking Age smallpox might thus have been a mild disease, possibly explaining the lack of historical records on smallpox epidemics during that time period.
Monkey pox epidemics in Africa
One might ask what is the purpose of reconstructing the natural history of a viral disease – even if it was one of the greatest killers of mankind - whose agent has now been eradicated. Isn’t it just of historical but not of biological interest? The recent worldwide surge of monkeypox disease is a striking reminder that orthopox viruses, the taxonomic group to which VARV and monkeypox virus (MPXV) belong, are still good for bad surprises.
MPXV is not VARV. Initially isolated from a monkey (due to its wide host range, the natural host is still unknown), its veterinary importance is limited. Research focus on monkeypox disease (mpox) was initially motivated by public health efforts to identify residual cases of smallpox-like disease during and after the final phase of smallpox eradication program. In the 1970s about 50 cases of smallpox-like disease were identified mostly in Central Africa. All cases were MPXV infections and epidemiological links to animal exposure (potentially from bushmeat) was revealed. Surveys conducted in the 1980s and 1990s in the Democratic Republic of Congo identified several hundred cases of mpox, mostly in children. Young children showed substantial mortality (CFR of 15%). Distinct from ordinary smallpox, an enlargement of the lymph nodes was characteristic for mpox. Squirrels and Cricetomys rats caught in the neighborhood of the cases showed antibodies against MPXV. A new picture for mpox disease emerged in a 2017 epidemic in Nigeria where young adults and not children were affected. Most patients had an HIV-1 coinfection. Currently two types of mpox disease are differentiated: one in Central and another in Western Africa. These two forms are associated with two distinct clades of MPXV; the Western African form is characterised by a lower CFR. Both forms of mpox disease in Africa received only modest research activities since only rare cases of export of mpox into the industrial world were documented. In 2003 a few cases of mpox disease were reported in the US, linked to pet animal trade from West Africa, where notably Cricetomys rats infected with the West African MPXV clade, were involved.
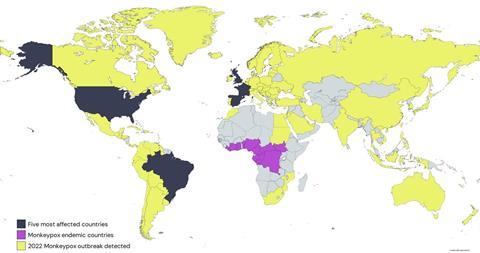
The worldwide monkeypox epidemic in 2022
The situation changed dramatically in 2022 with a worldwide epidemic of mpox, infecting more than 80’000 people but displaying a low case fatality rate. The epidemic is characterised by a number of particularities. Epidemiologically, nearly exclusively men having sex with men (MSM) were affected. Lesions were not numerous and limited to the anogenital and oral region. Local pain (proctitis, difficulty swallowing) were the principle clinical signs. Except for a few cases (health care workers, tattoo parlor visitors), no spread of mpox into the general population was observed. The epidemic showed a sharp rise period, followed by an equally sharp decline period with few new infections currently occurring. The viral isolates are genetically derived from the West African MPXV clade, but they showed an unusually high number of mutations. Two research groups attributed these mutations to a host enzyme of the innate immune system modifying the DNA of the virus. Disturbingly, the perhaps failed effort of the DNA base deaminase activity from the innate immune system to inactivate an invading virus might have provided a poxvirus an accelerated evolution, potentially creating new transmission pathways for this virus.
Conclusions
Fortunately, this mpox epidemic did not become a pandemic, by remaining restricted to a subgroup of the population and quickly running it course. However, several important warning lessons could be deduced from this epidemic. Even for a viral infection with a reproduction number R0 below 1 (i.e. below the maintenance level), as with MPXV in an unvaccinated population, an epidemic spread can occur in a highly connected network of people exploiting particular transmission routes. At the same time, quick transmission of public health messages in the highly connected MSM community might also have curtailed the epidemic. Due to limited public health resources in Africa, mpox remained a neglected infectious disease after the smallpox eradication phase. The intrinsic dynamic of a viral infection and the globalisation of the modern world showed how quickly a neglected infection from Africa can become a global concern, calling for more research on neglected tropical infectious diseases whose agents are just waiting for an introduction to populations outside their normal ecological range.
Harald Brüssow’s recent paper on the pandemic potential of poxviruseses is currently in press in Microbial Biotechnology, and can be read here.





No comments yet