A team of scientists may have solved one of the biggest hurdles standing in the way of synthetic biology - the difficulty of transferring the resulting large DNA molecules into bacterial host cells.
The researchers from Toyo University in Japan, compared physical methods of transferring DNA with DNA transfer based on bacterial genetic systems, the latter of which they say is subject to many restrictions in order to prevent foreign DNA invasion.
Their paper, ‘Approaches for introducing large DNA molecules into bacterial cells’ appears in the Journal of Applied Microbiology, a Applied Microbiology International publication.
Creating novel organisms
“In the near future, bacterial genome design will be performed on the computer, and the designed DNA will be synthesized using a DNA synthesizer. A novel organism will be generated with a defined genome,” corresponding author Dr Hiromi Nishida explained.
2Host-vector-mediated transformation of large DNA molecules of bacterial chromosome size requires repeated operations, which is time-consuming, labor-intensive, and costly. Additionally, the host bacterial species used in the repeated transfer of large DNA molecules are limited (Itaya et al., 2005; Kuroki et al., 2007; Gibson et al., 2008; Gibson et al., 2010; Hutchison et al., 2016).
“In contrast, liposome-mediated and microinjection-mediated transfers, which are useful for transformation using any bacterial genomes and host cells, can be performed by a single operation.”
Synthetic biology
Engineering of the bacterial genome plays a key role in systems biology and synthetic biology, Dr Nishida said.
“Genetic engineering of the bacterial genome involves the design and synthesis of large DNA molecules. However, functional studies of the designed and synthesized large DNA molecules are lagging,” he said.
“The development of DNA synthesizers has enabled synthetic studies of large DNA molecules (Venter et al. 2022). It will be possible to synthesize large DNA molecules of bacterial chromosome size (>1 Mbp) using DNA synthesizers.
“However, functional studies of designed and synthesized large DNA molecules have stagnated due to a lack of effective methods for transferring large DNA molecules into bacterial cells.
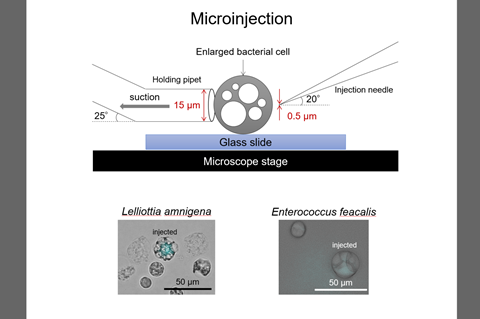
“It is necessary to establish experimental methods that can clarify how bacterial host cells recognize the transferred large DNA molecules of bacterial chromosome size. We have developed a method for transferring large DNA molecules into enlarged bacterial protoplasts using a micromanipulator.”
Bacterial protoplasts
In a study published last year, the team revealed how they had succeeded in transferring heterologous bacterial genomes into enlarged bacterial protoplasts using a micromanipulator (Takahashi & Nishida 2022).
“Two major methods can be used for transferring large DNA molecules of bacterial chromosome size into bacterial cells: transformation mediated by liposomes or by microinjection,” Dr Nishida said.
“These methods for transferring large DNA molecules into bacterial cells through a single operation will contribute to both fundamental and applied research in microbial genome science.
“In this review, we focused on methods for transferring large DNA molecules of the size of a bacterial chromosome, which are not feasible by DNA vector-dependent methods. DNA transfer based on bacterial genetic systems has many restrictions in order to prevent foreign DNA invasion. In contrast, DNA transfer by physical manipulation, such as microinjection, has few such restrictions.”
DNA transfer methods
Dr Nishida said: ”There are two methods of DNA transfer by physical manipulation: transformation mediated by liposomes or by microinjection. We contributed to develop a method of transformation by microinjection.
“Transformations mediated by liposomes and by microinjection require cell wall-deficient bacterial cells, L-form, protoplast, or spheroplast. The lipid composition and size of the cells should be considered before the transformation.
“In the liposome-mediated transformation, the liposome membrane and the plasma/outer membrane of the bacteria host cells should be fused. For that, the lipid composition of liposomes and the conditions at the fusion should be considered.
“In the microinjection-mediated transformation, the cell wall-deficient cells should be enlarged and their membranes should have a physical strength enough to withstand penetration of the microneedle.
Generating protoplasts
In order to transfer large DNA molecules into bacterial protoplasts using a micromanipulator, the scientists generated enlarged bacterial protoplasts in marine broth, considering the factors (Nishida 2020).
“We succeeded in microinjection of a blue fluorescent protein into enlarged bacterial protoplasts (Takahashi et al. 2020). After that, we succeeded in transferring heterologous bacterial genomes into enlarged bacterial protoplasts (Takahashi & Nishida 2022),” Dr Nishida said.
On what needs to be examined in the future to build on these findings, Dr Nishida said the large target DNA molecules of bacterial chromosome size should have been maintained during the transformation experiment.
“DNA methylation should also be considered as the chromosome DNA of the host cell is generally methylated at adenine and/or cytosine,” he said.
Critical to success
“In the liposome-mediated transfer, large DNA molecules must be encapsulated into the liposome, and the solution in the liposome should be considered. In the microinjection-mediated transfer, the buffer composition and the flow rate of injection are critical for success.
“Although novobiocin can control protoplast enlargement due to the inhibition of DNA replication (Kami et al., 2019; Tsuchikado et al., 2020), the enlarged cell cannot be made smaller at the moment. In many cases, the cell wall-deficient cells should revert to the cell wall cells that can divide and grow.
“The L-form cells can divide, but protoplasts/spheroplasts cannot. The protoplasts/spheroplasts of ≥15 µm in diameter did not revert to dividing cells even if the cell wall synthesis inhibitor was removed.
“Thus, we should establish the mechanism for reverting from enlarged protoplasts/spheroplasts to normal dividing cells, which re-synthesize the cell wall and maintain it. In addition, we should conduct studies on the transfer of large DNA molecules of bacterial chromosome size into L-form cells.”
Dr. Nishida paid tribute to Dr Isamu Yabe who originally taught them the method of bacterial enlargement (Kuroda et al. 1998). Dr. Yabe’s group have used the enlarged bacterial cell for patch clamp analysis.
‘Approaches for introducing large DNA molecules into bacterial cells’ appears in the Journal of Applied Microbiology.







No comments yet