Scientists have engineered lab-grown lung epithelial cells to light up red in colour once they launch an immune response, creating a tool that can be used to screen for drugs that can help treat COVID-19 and other emerging infections.
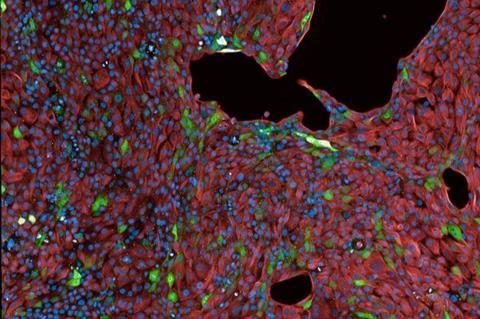
When we breathe, the SARS-CoV-2 virus enters the body through the cells of the upper respiratory tract. Sensing an invader, the epithelial cells, which are the first line of defence, mount an immediate and broad “innate” defense and trigger an alarm. The immune system then launches a cascade of vigorous, targeted attacks.
Understanding how these cells respond to infection and modulate their response pharmacologically is of paramount importance in developing effective treatments for viral diseases like COVID-19. The findings of this study, by Dr. Gaetano Gargiulo, head of the Molecular Oncology lab in the Max Delbrück Center, are reported in Science Advances.
Pandemic switch
This tool was originally developed in the Gargiulo lab to study cancer. But during the pandemic, the scientists decided to trial it in virus-infected cells.
“Our team decided to create this tool to understand and fight viral infections in the spirit of doing our part during this pandemic” says Gargiulo, senior author of the study. “It may be possible to rapidly address emerging pandemics in the future by tailoring our tool to recognize novel viral strains.”
The tool is called ’synthetic locus control region’ (sLCR) and it consists of a lab-generated segment of DNA that switches a fluorescent protein on or off depending on whether the cell is mounting an immune response. During an innate immune response, the sLCR is turned on and it makes a protein that glows red when observed under a fluorescence microscope, telling scientists that the cell is aware of being infected and also how strongly it is fighting back.
Unique DNA sequences
The scientists constructed an sLCR containing several unique DNA sequences which they had predicted would be active during a SARS-CoV-2 infection based on other studies. They inserted the sLCRs into epithelial cells grown in a petri dish, which could then be infected with the SARS-CoV-2 virus.
The cells glowed red once the innate immunity was activated by the infection or surrogate biochemical cues, and they were visualized using fluorescence microscopy.
“The most exciting moment was when we saw that the infection with different strains of the live virus actually triggers the colour coding,” says Ben Jiang, a graduate student at the Gargiulo lab and co-first author of the study.
Experiments with the live viral particles were possible thanks to the inter-Helmholtz collaboration between the Gargiulo lab and the group led by Luka Cicin-Sain at the Helmholtz-Zentrum für Infektionsforschung (HZI) in Braunschweig.
Finding new treatments
Such a simple read out enabled the scientists to look for drugs that inhibited or enhanced cells’ responses to the virus. They found that cells treated with some rheumatoid arthritis drugs did not glow red, suggesting the drug blocked the immune response. When the cells were treated with certain chemotherapy drugs, the cells glowed more intensely, suggesting the drug enhanced the immune response.
The opposing effects may prove useful at different stages of COVID-19. At the start, a drug that provokes a strong immune response could help battle the virus. But later on in the disease, a prolonged response could worsen the pathology.
“With a tool like this, one can identify compounds to strengthen or weaken the epithelial immune response, both of which can be useful depending on the disease stage and symptoms,” Jiang says.
Alert signal
In particular, the discovery that DNA damaging agents can enhance the alert signal from epithelial cells supports low-dose radiotherapy as a potential treatment for viral infections including COVID-19. This was tested during the pandemic, but needs precise dosage and timing, says Gargiulo.
Though this study was done in cell cultures, the drugs identified have been studied in clinical trials for COVID-19 by other groups. Hence, this tool could be used to screen drugs in bulk to find novel combinations and drugs that can be further trialled to see if they are effective in people. Moreover, the technology could be easily applied to more sophisticated disease models, such as organoids or mice, says the other co-first author Matthias Schmitt.
“The same approach can be readily re-purposed to target other viral infections, such as the emerging threat of Dengue and Zika viruses, and the technology is accessible to labs around the world to find drugs to combat emerging infectious diseases timely,” says Gargiulo.





No comments yet