When bacteria build communities, they cooperate and share nutrients across generations. Researchers at the University of Basel have been able to demonstrate this for the first time using a newly developed method. This innovative technique enables the tracking of gene expression during the development of bacterial communities over space and time.

In nature, bacteria usually live in communities. They collectively colonize our gut, also known as the gut microbiome, or form biofilms such as dental plaque. Living in communities provides many advantages to the individual microbes. They are more resilient against adverse environmental conditions, conquer new territories and benefit from each other.
Analyzing microbial communities
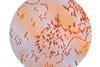
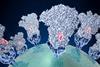
The development of bacterial communities is a highly complex process where bacteria form intricate three-dimensional structures. In their latest study published in Nature Microbiology, the team led by Professor Knut Drescher from the Biozentrum of the University of Basel has investigated the development of bacterial swarm communities in detail.
They achieved a methodological breakthrough enabling them to simultaneously measure gene expression and image the behaviour of individual cells in microbial communities in space and time.
“We used Bacillus subtilis as a model organism. This ubiquitous bacterium is also found in our intestinal flora. We have revealed that these bacteria, which live in communities, cooperate and interact with each other across generations,” explains Prof Knut Drescher, head of the study. “Earlier generations deposit metabolites for later generations.”
They also identified different subpopulations within a bacterial swarm, which produce and consume different metabolites. Some of the metabolites secreted by one subpopulation become the food for other subpopulations that emerge later during swarm development.
Distribution of tasks
The researchers combined state-of-the-art adaptive microscopy, gene expression analyses, metabolite analyses, and robotic sampling.
Using this innovative approach, the researchers have been able to simultaneously examine gene expression and bacterial behavior at precisely defined locations and specific times as well as to identify the metabolites secreted by the bacteria. The bacterial swarm could thus be divided into three major regions: the swarm front, the intermediate region and the swarm center. However, the three regions display gradual transitions.
“Depending on the region, the bacteria differ in appearance, characteristics and behavior. While they are mostly motile at the edges, the bacteria in the center form long non-motile threads, resulting in a 3D biofilm. One reason is the varying availability of space and resources,” explains first author Hannah Jeckel. “The spatial distribution of bacteria with distinct behavior enables the community to expand but also to hide in a protective biofilm.” This process appears to be a widespread strategy in bacterial communities and is crucial for their survival.
Complex dynamics
This study illustrates the complexity and dynamics within bacterial communities and reveals cooperative interactions among individual bacteria - in favor of the community. The spatial and temporal effects thus play a central role in the development and establishment of microbial communities.
A milestone of this work is the development of a pioneering technique that enabled the researchers to acquire comprehensive spatiotemporal data of a multicellular process at a resolution never before achieved in any other biological system.



No comments yet