Tackling worldwide disease burden requires, in part, the development of new medicines. Although medical advances have led to a decrease in mortality for many diseases (e.g. infectious diseases, cardiovascular disease), there is still a long way to go for rare diseases, metastatic cancers, diseases affecting the central nervous system and the ever-increasing infections caused by multi-drug resistant bacteria.
So, the question is how will we source the next generation of drugs? There won’t be a one size fits all solution, but many have acknowledged the important role of natural products (NPs) for future drug discovery efforts.
In the broadest sense, NPs are compounds or substances produced mainly by bacteria, fungi or plants known to be very diverse in chemical structures and biological activities. Beyond this, there is real no consensus on what constitutes an NP. Although, NPs have also been described as products of secondary metabolism – biological processes not necessary for growth or survival. This is the description used for the framing of this article. NPs are critical to how an organism interacts with its environment. The first medicines came from plant extracts, which contain a mixture of NPs.
NPs as therapeutic drugs
In an in-depth analysis of all drugs approved from 1981-2019, 46% of these were natural products, their derivatives, mimics or mixtures made from them. As such, NPs have made an important contribution to the establishment of contemporary drugs. Clinically revolutionary NP-sourced drugs include penicillin, morphine and paclitaxel. Additionally, the best-selling drug in the last 25 years is atorvastatin, a microbial NP derivative with total sales of $94.67 billion USD.
Historically, the contribution of NPs to the drug space was even larger. However, throughout the 1990s pharma shifted away from NPs and towards combinatorial chemistry – synthesis of new compounds by repetitive linkage of chemical groups - for the next generation of drug leads. The often-challenging process of NP isolation along with rediscovery of many NPs with similar modes of action led to this shift. Funding was diverted from the discovery and characterization of NPs. As such, many of the NP-based drugs approved today are based on the tweaking of NPs discovered decades ago. For example, vancomycin, discovered in 1957, is an antibacterial natural product that has been used to treat infections. Compounds telavancin (approved in 2009), dalbavancin (approved in 2014), and oritavancin (approved in 2014) were all derived from vancomycin and are also used to treat skin infections.
The need to reinstate NP-based drug discovery programs has been voiced by many. NP’s unmatched structural diversity, wide range of bioactivities and particularly, the higher likelihood of NPs modulating both other compounds and biological pathways compared to synthetic compounds make them interesting drug leads.

Biases in NP discovery
Most of the natural products characterized today, and in turn, those NPs successfully delivered to the drug market have been derived from analysing soil microbiomes, particularly bacteria belonging to the Actinomycetaceae family. Moreover, a great bulk of all NPs discovered so far have been the result of culturing efforts. The general workflow entails (1) assaying novel organisms in soil microbiomes, (2) culturing the organism and detecting the types of NPs it produces and (3) screening the isolated natural product for clinically relevant biological activities.
The bias in the methods used as well as the organisms studied for NP discovery means there is still great potential for the discovery of novel NPs with different biological activities by focusing on (1) poorly studied and cryptic environments (e.g. marine); and (2) the NP repertoire of microbial dark matter – NPs produced by organisms that have not been cultured before.
Given that many microbes are still to this day either unculturable or challenging to culture, it has been postulated that many of the next generation of natural products will be abstracted from genetic information made available by sequencing efforts. Metagenomic sequencing – bulk sequencing of a community of multiple organisms – enables scientists to analyze the genetic information harbored within an entire community and in turn, explore natural products produced by unculturable taxa.
The human microbiome and the NPs harboured within
The human gut microbiome is composed of trillions of microorganisms that aid in digestion, protect us against pathogens and regulate our immune system. The disruption and impoverishment of its native flora has been implicated in a plethora of human diseases. Although the human microbiome has an evident role in human health, the mechanisms by which the microbiome exerts its influence on health are unknown.
Nearly a decade ago, scientists observed that the human microbiome encodes the biosynthetic machinery for a plethora of unknown NPs with poorly understood biological roles. Understanding the roles that NPs produced by the human microbiome play can be the key to understanding how the microbiome influences health and as such, open doors to new therapeutic avenues. For example, NPs harmful for human health could be inhibited, whilst NPs with antibiotic activity may be administered to eradicate pathogens.
Genome mining, a bioinformatic and computationally reliant approach, refers to the search for genes involved in the biosynthesis of natural products. In the case of bacteria and fungi, biosynthetic genes are typically co-located in the genome and core enzymes are highly conserved facilitating the identification of gene clusters that encode NPs. Thus, using sequencing data alone one can predict which compound classes an organism has the capacity to produce.
Genome mining approaches have been applied to human microbiome metagenomes. This revealed that the biosynthetic repertoire of the human microbiome includes over 3,000 NP-encoding gene clusters. Additionally, a class of NPs, the ribosomally synthesized and post-translationally modified peptides (RiPPs) were found to be widely distributed across human microbiome body sites and relatively more abundant in human microbiomes compared to environmental ones. Three classes of RiPPs, the lantibiotics, thiopeptides and microcins were particularly abundant in human microbiomes. These NP classes can have antibacterial activity against both gram-positive and gram-negative bacteria. This has been explained through RiPPs’ involvement in the mediation of intra- and interspecies competition as well as niche colonization.
Efforts to characterize NPs flagged by genome mining approaches have yielded promising results. One such example has been the discovery of lactocillin, a thiopeptide antibiotic produced by Lactobacillus gasseri which has antibacterial activities against a range of gram-positive vaginal pathogens. Additionally, in another study, scientists found that when one of 21 NP gene clusters were identified in the human vaginal microbiome, Lactobacillus iners was absent. Clinically, this is an important observation as the presence of Lactobacillus iners has been linked to bacterial vaginosis infection.
Research efforts have also focused on characterizing a class of NPs, non-ribosomal peptides (NRPs) within human microbiomes. Though a class of NPs relatively impoverished in human microbiomes, large families of NRPs reside exclusively in the gut. Experimental characterization of NRPs produced by the human microbiome has been conducted. Previously, a scientific team (1) screened the genomes of human-associated bacteria for NRPs, (2) chemically synthesized these compounds and (3) went on to check if these compounds had any inhibitory effects against a set of both pathogenic and non-pathogenic bacteria. This led to the discovery of compounds now termed humimycins.
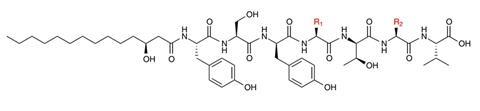
These antibiotics can disarm and inhibit methicillin-resistant Staphylococcus aureus (MRSA). Furthermore, a mouse study demonstrated that the administration of humimycins improved the survival outcomes of MRSA-positive mice. This is a remarkable milestone. MRSA is considered a high-priority pathogen that causes significant morbidity and mortality worldwide. MRSA positive patients have a mortality ranging from 20 to 50%.
Knowledge about the repertoire of NPs harbored within human microbiomes and their roles is still limited. Bridging this gap is essential to understanding the shaping of the human microbiome and its effects on health. The discovery of lactocillin and humimycins from human microbiome genome mining efforts showcase the therapeutic potential of human microbiome NPs. This represents only the tip of the iceberg of potential health-regulating compounds extrapolated from the human microbiome.








No comments yet