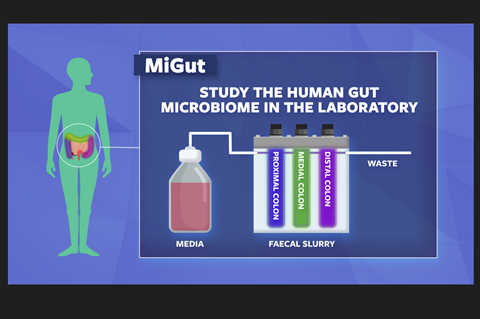
A benchtop model of the human gut (MiGut) has been developed to allow the interaction of drugs, nutrition, prebiotics, and live biotherapeutics within the gut microbiome to be studied in greater depth than ever before.
The work by a multidisciplinary team at the University of Leeds is outlined in a paper, ‘MiGut: a scalable in vitro platform for simulating the human gut microbiome – development, validation, and simulation of antibiotic induced dysbiosis’, which has been accepted by Microbial Biotechnology, an Applied Microbiology International publication.
Lead author William Davis Birch said the human gut microbiome (HGM) is inextricably linked with human health and disease and there is a growing need to understand how to modulate the HGM as a means of improving human health.
Useful tool
“Laboratory based models, where a live microbiome derived from human faecal samples is grown and sustained, offer a useful tool for studying the HGM. With these models, conditions such as pH, temperature, nutrient composition, and retention time can be precisely controlled in order to closely match the human colonic environment,” he said.
“Samples can be easily taken from each of the vessels simulating the different regions of the colon – something which cannot easily be done in animal models or humans.
“Laboratory based models allow longitudinal studies of novel therapies to be performed with far fewer ethical restrictions, which also helps reduce the use of animals in testing.
“Whilst traditional in vitro fermentation models have proved extremely useful, they are complex to set up and run, requiring large amounts of lab space, staff time, and resources. This has posed practical limits on their scalability, limiting studies to a small number of test conditions.”
Scalable biorector platform
The Leeds team has addressed the limitations of gut model systems by developing MiGut – a scalable bioreactor platform for studying the HGM in vitro. This work is part of a fruitful collaboration between the School of Mechanical Engineering (Prof. Nikil Kapur, Prof. Peter Culmer, and William Davis Birch), the Faculty of Medicine and Health (Dr Ines Moura, Mr. Duncan Ewin, and Prof. Mark Wilcox), and the School of Food Science and Nutrition (Dr Anthony Buckley) in developing state of the art tools for microbiological studies.
Each MiGut model consists of three vessels that simulate the proximal, medial, and distal regions of the colon. Conditions such as pH, temperature, nutrient availability, retention time, and hypoxia are closely controlled to match those of the human colon, making MiGut reflective of the in vivo environment.
Each MiGut platform consists of four parallel models that can be individually controlled to run as replicates or to investigate different test conditions. The platform has been specially designed so that it is easy to set up and operate, meaning that many models can be run simultaneously.
Real-time data monitoring
“We’ve also implemented a real-time data monitoring platform that automatically receives data from MiGut, displaying it in real time so experiments can be closely monitored remotely,” Mr Davis Birch said.
“We validated the MiGut platform by running a demanding nine-week study alongside a much larger, traditional triple-stage gut model. This larger model has enabled the development of novel treatment regimens that informed current antibiotic prescribing guidelines and supported drug regulatory submissions but is complex to run.
“We inoculated all models with a human faecal slurry to allow for a complex microbiome to be cultured. After a two-week equilibration period, the models were exposed to a series of antibiotics to induce a microbial imbalance or a “dysbiosis”.
“They were then run for a further four weeks to allow for the long-term effects of antibiotic dosing to be studied.”
Microbial ecologies
The initial data showed that the MiGut platform was able to recapture the microbial ecologies of faecal samples from different human demographics; i.e. the ecology put into each model at the start is the same ecology that we see after a couple weeks.
“When we stressed the microbial populations with antibiotics, we saw great correlation between the dynamics of the microbial populations between all models tested,” Mr Davis Birch said.
“Interestingly, our data showed that the microbial populations did not return to their original state, highlighting the long-term effects that antibiotic use can have on the HGM, even after prescription has stopped,” he said.
“The results confirm that the MiGut platform, with its improved ease of use and scalable nature, recaptures the performance of the larger human gut model and that the models themselves are highly repeatable.”
Addressing bottlenecks
The MiGut platform addresses one of the main limitations of in vitro gut model systems – the bottlenecks related to their scalability.
Although this paper only describes the operation of a single MiGut platform, the team have performed other studies with 12 simultaneous models and have plans for further increasing this number.
“We have coupled this increased throughput with a semi-automated molecular analysis for microbial populations. This is a significant increase in throughput compared to other systems and means that it is now possible to use in vitro gut models for complex studies with multiple variables and biological replicates,” Mr Davis Birch said.
“As research into the HGM continues to expand, researchers and industry will have an increasing need for models that are reliable, scalable, and can generate clinically reflective data. MiGut covers all of these requirements.
“Using these models, it is possible to study the HGM in ever-increasing detail, which will help us understand the underlying mechanisms that link it with human health and disease.”
Developing MiGut
The University of Leeds team is engaged in ongoing projects to develop the MiGut platform further and more accurately simulate the human colonic environment.
For example, they are interested in being able to reliably grow and sample the mucosal microbiota, which are known to be compositionally and functionally different to the microbes in the lumen.
“We are also modifying MiGut to be able to simulate the porcine hindgut, a system called RoboHog, which will allow us to study the pig microbiome to simulate the nutrient digestion and effect of novel therapeutics,” Dr Anthony Buckley said.
“We also have plans to expand our laboratory capacity, which will help to support wider interactions with academic and industry partners and will place the University of Leeds as a leading centre for in vitro microbiome research.
Host-microbe interactions
“As researchers continue to move away from using animal models, it will become increasingly important to be able to simulate the host-microbe interactions in vitro. Microbial fermentation models such as MiGut do not currently have this capacity.
“However, as gut-on-a-chip technology continues to improve, there will be increasing opportunities to study these interactions with the MiGut platform.”
This work has been supported by funding from the University of Leeds, National Institute for Health and Care Research (NIHR), and recently, Biotechnology and Biological Sciences Research Council (BBSRC).
‘MiGut: a scalable in vitro platform for simulating the human gut microbiome – development, validation, and simulation of antibiotic induced dysbiosis’ appears in Microbial Biotechnology.
Topics
- Agriculture
- Anthony Buckley
- Applied Microbiology International
- BBSRC
- Community
- Duncan Ewin
- Future Technologies
- Future Technologies
- Gut Microbiome
- Ines Moura
- Innovation News
- Mark Wilcox
- Microbiological Methods
- MiGut
- Modeling
- NIHR
- Nikil Kapur
- One Health
- Peter Culmer
- UK & Rest of Europe
- University of Leeds
- William Davis Birch





No comments yet