Human herpesviruses comprise the alpha, beta, and gamma subfamilies and are a widely prevalent group of DNA-enveloped viruses, capable of establishing lifelong latent infections in humans and causing various diseases.
Among them, herpes simplex virus (HSV) belongs to the alpha herpesvirus group and infects a wide population, causing symptoms like oral or genital herpes.
As an enveloped virus, HSV possesses a series of glycoproteins involved in virus recognition, adhesion, and infection processes. Among these, gB serves as the viral fusion protein, mediating the fusion between the virus and host cell membranes, and it is highly conserved across the entire herpesvirus family.
Therefore, gB represents an ideal antiviral drug or antibody target. However, the lack of structural information on neutralizing epitopes for HSV has hindered the design of antibody drugs and vaccine targets.
Investigating the target
The research team successfully purified the HSV-1 gB/D48 Fab complex using the 293F expression system. They validated through in vitro binding and neutralization assays that the D48 antibody efficiently neutralized HSV-1 virus infection by binding to gB (Fig. 1B/1C).
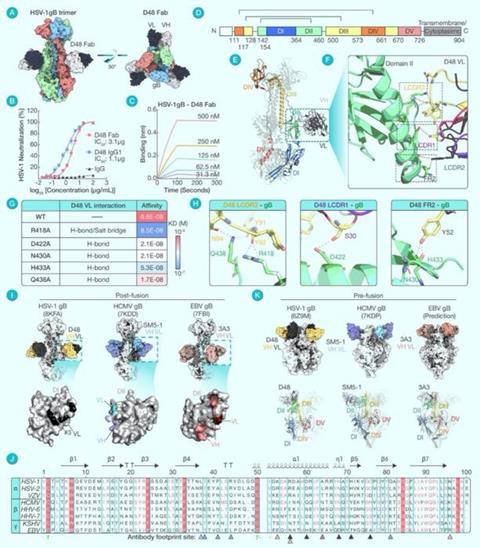
Subsequently, they resolved the structure of HSV-1 gB bound to D48 at a resolution of 3.04 Å using cryo-electron microscopy (Fig. 1A). The structure revealed that HSV-1 gB consists of five distinct domains (Fig. 1D), with D48 Fab primarily binding to the DII structural domain (Fig. 1E/1F).
Specifically, residues R418, D422, N430, H433, and Q438 from the LCDR1, LCDR3, and FR2 of the D48 light chain were found to interact polarly with DII (Fig. 1H), and alanine mutations at these key residues significantly reduced the interaction between HSV-1 gB and D48 (Fig. 1G).
Conserved nature
Upon obtaining high-resolution complex structures of neutralizing antibodies bound to HSV-1 gB, the research team performed a parallel comparison of the neutralizing epitopes of HCMV and EBV gB. They found that the neutralizing antibodies SM5-1 for HCMV and 3A3 for EBV also bind to the DII structural domain of gB (Fig. 1I). Further, they discovered the cross-herpesvirus conserved nature of these domains.
The footprints of the antibodies’ binding were not found to overlap with conserved residues, suggesting the likely existence of antibodies with broad-spectrum neutralizing activity against herpesviruses (Fig. 1J).
Additionally, the team compared the structures of the DII domains of HSV, HCMV, and EBV gBs in complex with antibodies to their corresponding pre-fusion conformations. This comparison yielded recombinant structures showing that the DII domain in the pre-fusion conformation retains the ability to bind neutralizing antibodies, indicating a conservative antigenic epitope presentation in both pre-fusion and post-fusion states (Fig. 1K).
In conclusion, this study, published in hLife, proposes that the gB DII domain within the herpesvirus family constitutes a broadly conserved neutralizing epitope structural domain. It highlights the exposure of antigenic epitopes in both pre-fusion and post-fusion conformations, providing crucial insights for the development of broad-spectrum drugs and vaccines against herpesviruses.
Key:
(A) Cryo-EM structure overview of HSV-1 gB in complex with D48 Fab. The side view and top view were presented. The HSV-1 gB trimer and D48 Fab VH/VL were colored differently.
(B) Neutralization assay of D48 Fab, D48 IgG1 and control IgG to HSV-1 infection of Vero cells. The neutralization (%) was calculated by [(mean plaque number of blank neutralization - plaque number of certain antibody neutralization at certain concentration) / mean plaque number of blank neutralization] * 100(%). Half maximal inhibitory concentrations (IC50) were calculated by non-linear regression for each antibody. Data points were shown as the mean ± SEM from three replicate wells.
(C) Biolayer interferometry (BLI) kinetic assay to D48 Fab binding to HSV-1 gB. Different concentrations were marked beside the corresponding curve in different colors. Kinetic data from one experiment was shown.
(D) Sequence and domain segmentation of HSV-1 gB. Domains were highlighted in different colors and interval residue positions were marked.
(E) Ribbon structure for a monomer of HSV-1 gB in complex with D48 Fab. The gB domain colors were consistent with (D).
(F) Zoomed-in view shows polar interaction diagram for HSV-1 gB DII domain with D48 Fab VL. The interaction residues were shown as sticks. The three CDRs and FR2 were colored differently and zoom views for LCDR1, LCDR3, and FR2 were outlined.
(G) Details for HSV-1 gB polar interaction residues with D48 Fab and the residue-mutant gB proteins affinity with D48 Fab measured by BLI. The listed polar interaction included hydrogen bonds (H-bonds) and salt bridges. The affinity was represented by the KD reported by BLI assays on wildtype (WT) or alanine-mutant HSV-1 gB binding to D48 Fab. Deeper red meant stronger affinity and deeper blue meant weaker.
(H) Detailed interaction diagram for HSV-1 gB polar interaction with D48 Fab in separate zoom view for LCDR3, LCDR1, and FR2. Key interacting residues of D48 or HSV-1 gB were marked in color consistently to (F).
(I) Comparison of architecture and antibody binding footprint of HSV-1, HCMV, and EBV post-fusion gB in complex with its DII-specific antibody. The VH and VL of D48, SM5-1, and 3A3 were colored differently. The antibody footprints on the DII domain were consistently colored by VH or VL.
(J) Sequence alignment of eight different gB DII domains across α, β, and γ herpesviruses. The secondary structure annotation was generated in the template of HSV-1 gB. Identical residues are shown as white symbols on red background, and similar residues are shown as red symbols. The exact residues of antibody footprint on DII domains were marked as arrows and colored by the corresponding binding antibody VH or VL in (I).
(K) Comparison of the architecture of HSV-1, HCMV, and EBV pre-fusion gB in complex with its DII-specific antibody generated by pre-post DII domain alignment. The VH and VL of D48, SM5-1, and 3A3 were colored differently as (I).





No comments yet