Researchers who scoured deep sea sponges in search of novel antimicrobial compounds have discovered several bacterial strains that are effective against a variety of pathogens.

The discoveries by a team at the University of Plymouth are outlined in ‘Whole genomes of deep-sea sponge-associated bacteria exhibit high novel natural product potential’, which is published by FEMS Microbes.
The study was led by Dr Poppy Hesketh-Best and Dr Matthew Koch, who through the period of their PhDs cultivated and screened hundreds of bacterial isolates. Dr Koch’s PhD was sponsored by Applied Microbiology International.
It investigated the potential of deep-sea bacteria isolated from sponges for producing novel antibacterial compounds and presents draft genomes of 12 isolates, including four potentially novel strains.
Antibiotic resistance
“The overuse and misuse of antibiotics has resulted in global antibiotic resistance, making it difficult to treat infections and safely perform many medical procedures,” Poppy Hesketh Best said.
“One strategy to combat this crisis is the discovery of new antibiotics or alternative antimicrobial compounds by exploring new environmental habitats, such as the deep sea. Environmental bacteria produce a variety of chemical compounds that may have antimicrobial properties which we can access when we grow and study these organisms,” she said.
The team carried out their study on glass sponges, a type of sponge found in deep water.
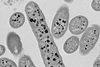
“We spent a long time growing as many bacteria as we could from these sponges, hoping to find some that might be able to produce antibiotics,” Dr Hesketh Best said.
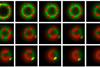
“To do this, we ground up the sponges with artificial seawater and put the diluted mixture onto agar plates, leaving us with a collection of bacterial samples that all had different levels of antibiotic production.
“However, the activity observed in the lab can be misleading, as environmental bacteria don’t always express their full range of products in lab conditions. To get a better understanding of their capabilities, we focused on sequencing the genomes of twelve of the most promising candidates.”
Potentially new species
While investigating genomes, the team found four potentially new species that couldn’t be identified just by looking at the 16S rRNA gene commonly used to identify bacteria.
“We also found unique genes that produce different compounds from what we’ve seen before, which is a promising sign we are exploring new chemical spaces,” Dr Matthew Koch said.
“Lastly, when we grew these bacteria on different nutrient sources, we found that they were effective against various pathogens. These three results together are a good indication that these new bacteria and their unique genes have the potential for producing a range of useful compounds, as shown by their different results when grown in the lab.”
Antibiotic discovery pipeline
The project is at an early stage in the antibiotic discovery pipeline, but is no less important.
“We now have these candidates in culture in the lab and their genomes on the computer - both will help direct more targeted exploration of the products these bacteria are capable of producing and currently produce,” Dr Hesketh Best said.
“The chemicals that we can observe from these organisms, especially those that are active against pathogens of concern such as Staphylococcus aureus, need to be chemically and structurally investigated.”
This study was supported and advised by Professor Mathew Upton, Professor Kerry Howell, Dr Phil Warburton, and Dr Grant January from the University of Plymouth.




No comments yet