Methanotrophs are microorganisms (bacteria or archaea) that feed on methane gas for their energy and growth. Back in 1906, N.L. Sohngen discovered the first known bacteria that could use methane, named Bacillus methanica.
A 600 million ton problem
In 1906, Sohngen suspected that these microorganisms might be helping to decrease the amount of methane in the air. This suspicion came about because even though a lot of methane was being released into the air from natural and human sources, the levels of methane in the air seemed lower than expected.
In 1970, Roger Whittenbury and his team found about 100 different methanotrophs using a method called ‘Plate Microscopy’. This discovery was a big deal, and sparked a field of research determined to understand these methane-consuming bacteria. However, we now know that most of the methane in the air is actually broken down by something else called hydroxyl radicals in the lower atmosphere. Only around 10% of methane in the environment is taken up by these methane-consuming bacteria, and they’re mostly found in natural habitats like water, soil, caves, and rice paddies.
Unfortunately, since the industrial age - and with more people on the planet - human activities have been making far more methane than we used to. Methane levels have gone up more than two and a half times since 1750, at a much faster rate than carbon dioxide. Methane is now the second biggest human-made greenhouse gas after carbon dioxide.
In 2017, almost 600 million tons of methane were released into the atmosphere, with around two-thirds coming from agriculture, and the rest from burning fossil fuels. Currently, the amount of methane in the air is about 1922 parts per billion (ppb). Even though methane is extremely good at trapping heat within the atmosphere (about 86 times more powerful than carbon dioxide), it’s still much less common than CO2.
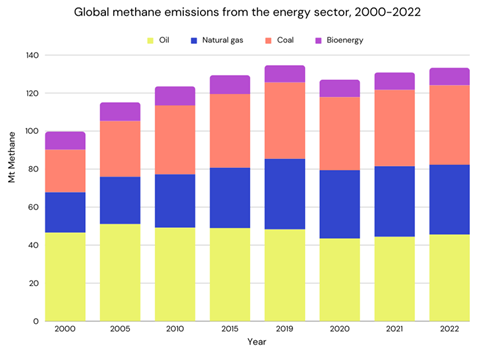
Capturing methane from the air is hard with today’s technology because there isn’t a lot of it around. So, scientists are exploring other ways to deal with it. Some particularly cool ideas involve using seaweed and special oils in cows’ food, which can help reduce the methane they release when they burp. Also, changing how rice fields are flooded can cut methane emissions, although it may increase another greenhouse gas, nitrous oxide.
But here’s where the microbes come in. Methane-consuming bacteria turn waste greenhouse gases into useful products. It’s a win-win situation!
Enter the methanotrophs
Methanotrophs are remarkably diverse which has led to several categories of classification over the past few decades. In broad terms, they are either aerobic or anaerobic, based on their need for oxygen to survive. Aerobic methanotrophs, which are the most well-studied and the only form utilised commercially in the biotechnology industry are Gram-negative bacteria, further divided into γ-proteobacteria, α-proteobacteria, and Verrucomicrobia. γ-proteobacteria, also classed as Type I, assimilate carbon through the ribulose monophosphate (RuMP) pathway, synthesize pyruvate as a key intermediate, and use glycogen as a main storage compound. First isolated from the Roman bath in the City of Bath, UK by Roger Whittenbury and colleagues, the best-known Methanotroph is Methylococcus capsulatus Bath - widely studied as a suitable candidate for making single cell protein (SCP).
The second significantly studied group of methanotrophs are α-proteobacteria (also classed as Type II) which assimilate carbon through the serine pathway, synthesize acetyl-coA as a key intermediate, and store reducing equivalents in the form of polyhydroxybutyrate (PHB). Methylocystis parvus OBBP is a particular example.
Verrucomicrobia, the last and least-studied group of aerobic methanotrophs assimilate carbon through the Calvin–Benson–Bassham (CBB) cycle and are also classed as Type IV. To date, there has been no reported use of this group for commercial biotechnological purposes, although they have remarkably interesting characteristics such as growth at extremely low pH and high temperatures. In November 2022, a species of actinobacteria closely related to Mycobacterium tuberculosis named Candidatus Mycobacterium methanotrophicum was reported to aerobically use methane as its sole source of carbon and energy. Carbon assimilation was achieved through RuMP similar to Type I γ-proteobacteria, although with an extremely slow doubling time, averaging 150-200 days.
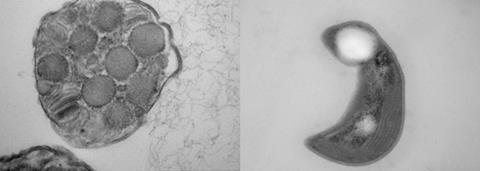
Anaerobic methanotrophs, first discovered in 1976, oxidize methane either through reverse-reaction of the first enzymatic step of methanogenesis, or by generating oxygen intracellularly in a process referred to as intra-aerobic oxidation, subsequently using the oxygen for methane oxidation. The former process, dominated by archaea, is possible through the Methyl-coenzyme M reductase (MCR) enzyme system similarly present in methanogens and tends to use manganese, sulfate, nitrate, iron and arsenate as electron acceptors. The latter, first reported in 2010 in Methylomirabilis oxyfera, converts two nitric oxide molecules to dinitrogen and oxygen using nitrite reductase. The oxygen is then used to oxidize methane.
This discovery was pivotal because it added to the list of ways biological oxygen was biosynthesized, in addition to photosynthesis, chlorate respiration, and the detoxification of reactive oxygen species.
Chemical methane oxidation
Methane, despite being known for its flammability, is actually not very reactive. This makes it tricky to activate and selectively transform its C-H bonds. For instance, turning methane into methanol chemically involves a complex process that starts with making syngas (a mix of CO and H2) from methane. This usually needs high temperatures, consuming massive amounts of energy for this expensive process. Plus, it’s hard to control because the resulting methanol is more reactive than the original methane, leading to the formation of unwanted products.
Scientists have tried different ways to improve this process over the years. Some have used strong chemicals like sulfuric acid or nitrous oxide to help convert methane, but these were too costly or caused corrosion problems. Recently, they’ve been developing special catalysts that help convert methane using regular oxygen at lower temperatures, sometimes as low as -50˚C. These catalysts, made with elements like iron or copper in a structure similar to those found in certain bacteria, have shown promising results in converting methane efficiently.
In September 2023, a study reported a new catalyst made of MoS2 (Molybdenum Disulfide) that could turn methane into useful compounds like methanol at just 25 °C. This catalyst achieved a good conversion rate of methane into these compounds without creating too much carbon dioxide. However, separating these different compounds afterward can be a challenge, and unlike natural enzymes found in certain bacteria, it’s not perfect – it can’t achieve 100% efficiency and selectivity.

Biological methane oxidation
Biological systems overcome methane inertness by using the methane monooxygenase (MMO) enzyme system. MMO and a homolog expressed by ammonia oxidizers, ammonia monooxygenase (AMO), are the only known natural catalysts that drive methane oxidation at room temperature. MMO is present either in soluble form (sMMO) which uses a diiron active site, or in a particulate membrane-bound form which possesses a catalytic copper centre (pMMO). Most methanotrophs studied possess the pMMO. A few possess the sMMO, some of which also have pMMO, thereby possessing both.
How it works:
MMO works by first activating O2 and inserting one atom into an inactivated C–H bond of methane. The process requires two reducing equivalents from NAD(P)H to split the O-O bond of O2, with one forming H2O while the other oxygen reacts with CH4 to form methanol - a reaction approaching 100% efficiency. MMO is very selective to methane as the substrate, meaning that overoxidation of methanol does not occur. Methanol is further oxidized to formaldehyde and formate in two-electron steps by other pathway enzymes to yield carbon dioxide (CO2). These reactions from formaldehyde to CO2 produce the reducing power for the initial oxidation of CH4 and cellular biosynthesis. The methanotrophs utilize some of the carbon at the oxidation to formaldehyde as cellular carbon for growth.
Apart from the CH4 oxidation properties that sMMO and pMMO share, both enzyme systems have striking structural, functional, and regulatory differences. The pMMO is expressed when the copper-to-biomass concentration in the growth media is relatively high. In most species, it comes in three subunits. For example in M. capsulatus Bath, it is constructed of pmoA, pmoB and pmoC. pmoA has a conserved gene region and is possessed by most methanotrophs, which has made it an important gene probe that indicates the presence of methanotrophs in the environment. The structure and function of the active sites of pMMO however, remain disputed. The main reason for this uncertainty is its unstable nature. Despite extensive efforts to stabilize the enzyme, the specific activity remains incredibly low and not highly reproducible.
Biotechnological applications
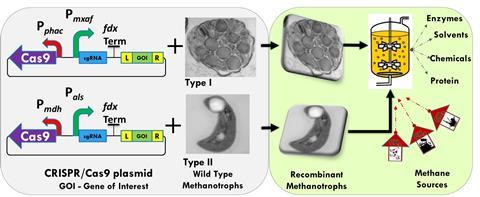
Over the last two decades, efforts have been made to develop the ways that methanotrophs can be utilized for metabolic engineering through the expression of heterologous genes, gene deletion, and overexpression of native genes. Among the very first protocols for the transfer of genetic material through conjugation was reported by J. W. Drozd et al. in 1980, where a P1 group plasmid was transferred into M. trichosporium Ob3B. Using these gene transfer tools, nitrogen-fixing mutants were generated in 1984 via conjugative transfer of a Tn5 plasmid. In 2008, electroporation was reported in a species of Methylocystis SC2. Several genome insertion and deletion technologies such as marker-exchange mutagenesis, the Cre-lox system, and the sacB-based deletion system have been the choice of genome editing technologies.
Since 2019, advanced genetic engineering tools such as CRISPR-based genome editing started to emerge. In early 2023, two publications demonstrating this further eased genome editing in methanotroph species. This is very crucial considering methanotrophs have the ability to convert waste greenhouse gases into valuable bioproducts. In the last three decades, several of these products have been studied providing insight into the opportunities that lie ahead.
Animal feed
Research into these products was predated by single cell protein (SCP). SCP refers to the entire microbial biomass, including the cell membrane and intracellular components of single or multi-species microorganisms grown using fermentation processes in a bioreactor. SCP has been shown to be a good protein source for animal feed. Several microbial organisms can be used as SCP but methanotrophs make good candidates as they tend to have higher protein content (up to 80% of Dry Cell Weight - DCW) when compared to yeast and fungi, as well as better digestibility than algae. Their favorable amino acid profile also makes them suitable ingredients for fish, pig, and chicken feed amongst others.

As far back as the 1960s SCP became the first methane-derived product obtained from methanotrophs. The project did not lead to long-term commercial success, as it could not compete with cheaper animal feeds (crops) that were available at the end of the 1980s. In 2022, a joint venture established between the feed supplier Adisseo and the US/UK biotechnology company Calysta gave rise to Calysseo, estimated to produce 20,000 tons of SCP per year using methane as feedstock from its production facility in Chongqing, China. The capacity is also projected to rise to 80,000 tonnes per annum in the near future. The likely strain used was the type I methanotroph Methylococcus capsulatus, shown to accumulate up to 75% protein per DCW.
There have also been reports of a mixed culture of M. capsulatus Bath producing higher protein content. UniBio is a company based in Denmark that uses a special method called continuous-flow microbial fermentation. With this method, they can produce about four kilograms of SCP in one cubic meter, every hour. This protein is used as animal feed and sold under the brand name UniProtein®. SCPs are becoming popular replacements for current livestock feed ingredients such as soy protein and fish meal for reasons including mainly lower carbon and water footprint. SCP has been shown to alleviate enteritis induced by soybean meal in sea bass. In other cases, such as in juvenile American eel, it did not show a significant difference in growth compared to fish meal but was reported to be more nutritional and cost-effective. One limitation of SCP is the presence of nucleic acids, particularly RNA, which leads to increased plasma levels of uric acid resulting in kidney stones and gout in humans. This has limited effect when added to animal feed, but its removal is essential if SCP were to become part of the human diet. The nucleic acid problem is usually managed by heat treatment of the cells during downstream processing.
Alternatives to plastic
Another important product methanotrophs produce is biodegradable plastic polymers called polyhydroxyalkanoates (PHAs). Polyhydroxybutyrate (PHB), a specific type of PHA is produced by most type II methanotrophs although synthesized by a wide range of microorganisms. They are biodegradable, biocompatible, and biorenewable, making them ideal for complementing and subsequently replacing synthetic fossil plastic. However, the expansion of PHB utilization has been restrained due to their high production cost.

Using waste methane as feedstock can reduce the cost of PHB by approximately 30-35%. This is crucial if PHB producers plan to take a market share of synthetic plastic, a market predicted to grow to up to 500 million tons by 2050. PHB is accumulated by methanotrophs under nitrate or ammonium starvation in growth culture media. Without optimization, about 40% DCW is achieved. However, up to 73% DCW has been achieved by a strain of Methylocystis hirsuta in media deficient in magnesium and phosphorus. Although PHB is the most commonly produced PHA, the material properties of pure PHB may not be ideal for many applications. PHB is relatively brittle, and its melting temperature is close to the temperature at which degradation begins, which creates a relatively narrow thermal processing window and can make polymer processing difficult. PHB copolymers have improved mechanical properties and have a wider thermal processing window due to a lower melting temperature.
Methanotrophs can be enabled to biosynthesize copolymers such as PHB-co-HV by adding one of the monomers, for example, valerate, in the growth media as co-substrate in addition to methane. PHB-co-HV has been accumulated at a concentration of 78% from Methylocystis sp. WRRC1 grown in copper-free mineral salt media with methane and a valerate as co-substrate. Methylosinus trichosporim has also been engineered to produce poly-3-hydroxybutyrate-co-4-hydroxybutyrate copolymer, opening the door for more desirable copolymers of PHAs. Mango materials in California produces pure PHB and biogas from wastewater treatment plants and landfills to make biodegradable plastic pellets referred to as YOPP.
The future of methanotroph research
Methanotrophs hold enormous potential for mitigating climate change, while simultaneously producing valuable products. To achieve this on a commercial scale, key challenges need to be addressed. This will involve optimizing the strains used for specific product biosynthesis, the fermentation technology that will support the growth of the strains on large scales, and finally, product separation and removal from cells where applicable.
Developing good fermentation technologies is crucial to the commercial success of any biotechnological product. This is because if correctly designed, bioreactors for growing methanotrophs at a commercial scale can solve the problem of poor methane and oxygen solubility in growth media, the fundamental challenge limiting the use of methanotrophs for biotechnological applications. Several bioreactor designs and technologies have been developed over the years, each with its advantage and prospects for promoting high gas-liquid mass transfer, as well as volumetric mass-transfer coefficient (KLa), thereby enhancing substrate consumption, cellular growth, and ultimately high product yield.

Finally, it is important to acquire the product from the bacterial cells after growth and production. This is where products such as SCP are advantageous considering the whole biomass is utilized and only needs to be dehydrated and then pelleted. Products such as PHB require the use of toxic chemicals for separation and processing which sometimes diminishes the environmental credentials of the product.
Regardless of the challenges that lie ahead, methanotrophs remain the only microbial workhorse that can harness the carbon and energy locked in methane gas.
You can read more about the work of Ying Zhang’s group at the University of Nottingham here.











No comments yet