The bacterium Pseudomonas aeruginosa adapts its biofilm form depending on the infectious site where it is found, potentially affecting antibiotic sensitivity, according to new research being presented at the Letters in Applied Microbiology ECS Research Symposium today.
Ayomikun Kade, of the University of Warwick, is presenting his research, entitled ‘Host-mimicking media impact on the biofilm biomass and composition of Pseudomonas aeruginosa’.
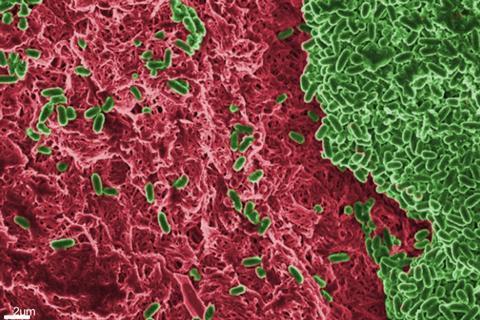
“We want to understand how the host environment influences the fundamental biofilm matrix of Pseudomonas aeruginosa, an important opportunistic pathogen at different infection sites. We discovered that Pseudomonas aeruginosa biofilm composition and matrix vary depending on the infectious site and this could impact antibiotic sensitivity,” he told The Microbiologist.
Varying biofilm matrix
“In my work, I have found that P. aeruginosa varies its biofilm matrix in media that mimic cystic fibrosis sputum, wound fluid and airway surface liquid versus standard lab medium. This supports other work carried out in my supervisors’ labs, which has found that P. aeruginosa changes its transcriptome and antibiotic tolerance in host-mimicking media versus standard lab media.
“These crucial findings stress the need to consider infectious sites as unique environments, and to adopt validated models and media that mimic infection environments. This will help to determine how well potential antibiofilm therapies in the drug discovery pipeline might work in a patient.”
A number of bacterial pathogens, including Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa, are challenging to treat and are listed by WHO as priority pathogens needing crucial drug intervention.
Planktonic or aggregating
These bacterial pathogens can exist in planktonic states or aggregate in a self-produced extracellular matrix to form a biofilm. Bacterial biofilms are responsible for 80% of chronic infections and are between 10 and 1000 times more resistant to host defences and conventional antibiotics.
“These pathogens are often opportunistic, able to form biofilms at more than one infectious site. For instance, P. aeruginosa biofilms are found on medical devices, in the lungs of people with cystic fibrosis, in endocarditis, and in wound infections,” Mr Kade said.
“Generally, studies of these pathogens in the laboratory use general-purpose media, which does not replicate the specific environments encountered by the organism in vivo. This often creates a scenario where an antibiotic active against a pathogen in vitro is inactive in vivo and vice versa.
“To avoid this scenario, there is a need to develop and adopt media that closely mimic the milieu at the infectious site, so that lab studies can produce results that better predict what happens in vivo.”
Mimicking the environment
Mr Kade said his supervisors’ labs often work with synthetic cystic fibrosis medium (SCFM) and synthetic wound fluid, to mimic the environment of the lungs of people with cystic fibrosis, and within chronic wounds, respectively.
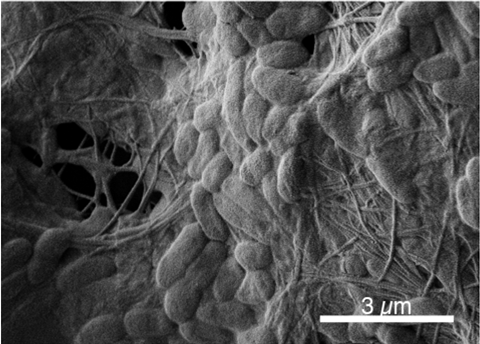
“Recently, colleagues in Freya Harrison’s lab – led by our post-doc Dean Walsh – have developed a new medium that mimics the airway mucus in intubated patients (synthetic ventilated airway mucus, SVAM). In the first year of my PhD, I have started to compare how P. aeruginosa and S. aureus form biofilms in SCFM and SVAM,” he said.
He did this by cultivating two laboratory strains of Pseudomonas aeruginosa (PAO1 and PA14) in these media, or in the standard rich lab medium lysogeny broth, in microtiter plates for 48 hours to achieve mature biofilm growth. The biofilm biomass was estimated through crystal violet staining.
By targeting the three major components of the biofilm matrix, viz eDNA, protein, and polysaccharide, through enzymatic degradation, Mr Kade was able to calculate the biofilm’s relative composition in the different media, and found that they form more biofilm in standard media than the different host-mimicking media which could be attributed to the nutritional content of the media.
Variations in media
“I also observed that polysaccharide was abundant in the biofilm matrix across different media but interestingly, there were significant variations in the amount of eDNA and protein in the different media. I also performed antimicrobial susceptibility testing using meropenem and colistin in the different host-mimicking media,” he said.
What he found surprising was the huge difference in biofilm composition and reaction to antibiotics by the same strain in different host-mimicking media.
“The findings from my PhD will have significant implications for understanding and addressing the problem of biofilm-associated infections caused by P. aeruginosa and other bacterial pathogens in real-world clinical settings,” he said.
“By using physiologically relevant media and models, we can more accurately represent biofilm formation and antibiotic tolerance in vivo. This research will help determine the right antibiotics for effective treatment and reduce the risk of antimicrobial resistance evolving in a chronically-infected patient.
“In drug development pipelines, using relevant media and models which mimic the specific host milieu found in a particular infection can help determine the potential efficacy of the drug in vivo.”
More advanced host-mimicking media
The next step will be to build and use more advanced host-mimicking media and models which incorporate host cells or tissues, immune components, and dynamic flow conditions to further simulate the physiological conditions encountered by bacteria in cystic fibrosis lungs, ventilator tubes and chronic wounds.
“This will allow me to better understand the biofilm of these pathogens and their interaction with host tissue, the spatial distribution of cells within a biofilm especially in polymicrobial infections and to explore the various components of the biofilm including the quorum sensing molecule at a higher resolution using state-of-the-art mass spectrometry.”
This study was led and supervised by Freya Harrison from the University of Warwick and Kim Hardie from the University of Nottingham. The BBSRC funded it through the Midlands Integrative Biosciences Training Partnership (MIBTP). The development of SVAM was led by Dean Walsh and aided by Jennifer Bevan.
Topics
- Antimicrobial Resistance
- Applied Microbiology International
- Ayomikun Kade
- Bacteria
- Biofilms
- colistin
- Community
- cystic fibrosis
- Dean Walsh
- Early Career Research
- Freya Harrison
- Jennifer Bevan
- Kim Hardie
- Meropenem
- One Health
- Pharmaceutical Microbiology
- Pseudomonas aeruginosa
- Research News
- synthetic cystic fibrosis medium
- synthetic wound fluid
- UK & Rest of Europe
- University of Nottingham
- University of Warwick




No comments yet