Megan Stenton reports back on her AMI-sponsored summer studentship which investigated the frequency of the SCCmec gene - a mobile gene element that houses the methicillin resistance gene - across members of the same species of Staphylococcus aureus.
Megan (22), from Bradford, completed her BSc in Biomedical Sciences at the University of Bradford this summer and has spent the summer working with Dr Sara Henderson at the University on her AMI Summer Studentship.

During my placement I was interested in how the mobile genetic element known as the Staphylococcal cassette chromosome mec (SCCmec) moves between different Staphylococcus aureus strains. This is important because it is this piece of DNA that causes the methicillin resistance in MRSA (Methicillin resistant Staphylococcus aureus).
While some people have looked at this before, the Bradford Infection Group have a group of MRSA strains collected at local hospitals which is interesting because of the variations in the local community. These include significant diversity in ethnic and socio-economic communities, and the rural and urban environments that the local hospitals provide care to.
During the placement I performed a range of proof of principle and fundamental experiments teaching me the fundamentals of experimental design. These included checking that our local Bradford MRSA collection were resistant to Oxacillin – we used this because it’s in the same class of antibiotics as methicillin but the results are easier to interpret. I also checked that the recipient strains were both oxacillin-sensitive and rifampicin-resistant and determined how I could isolate any strains that had collected the SCCmec and not confuse them with either the donor or the recipient.
Molecular profiling
In addition, I did some molecular profiling of the MRSA collection – the SCCmec in different MRSA strains is made up of different combinations of accessory genes which may play a role in its mobility.
This allowed me to learn another technique and to find a bit more out about our strains. For this I used a PCR screen with many primers at the same time known as a multiplex reaction.
Unfortunately, it didn’t work as expected so I had to split this into a couple of different reactions. In the first lane I had the primers for the mecA gene and the Type II, III, Iva, IVd and V, while in the second empty lane was Type I, IVb and IVc. I used an E. coli as a negative control which I knew shouldn’t react with my primers.
I got some nice bands which showed all my samples carried the gene for methicillin resistance (lower band), but the environment they were carried in was either a type III or V in our collection (upper band). This is interesting because some people have suggested Type IV to be most prevalent in the UK. However, as I only tested a very small sample of the collection, maybe I got lucky and only found the interesting strains.
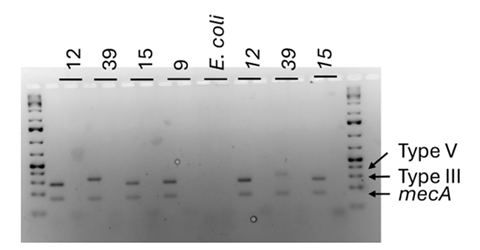
After confirming specific selection S. aureus NCTC 13297 was chosen as the recipient strain for the SCCmec, and MRSA 19, 15, 37, 39 and 9 were determined to be appropriate donor strains. Co-culture biofilms at different donor: recipient ratios (1:1, 1:10, 1:100) were produced and then spread plated onto dual antibiotic agar (Oxacillin and Rifampicin) to examine for any transformants. Transformants were identified in all donor: recipient pairs, even in those with different SCCmec types (type III and V, as determined by M-PCR).
However, quantification was more challenging with anything from 10-95000 colonies being obtained. This is a lot more than either me or my supervisor were expecting to obtain. I did observe that having more donors increased the number of double resistant colonies.
As rifampicin resistance can be caused by spontaneous mutations, it is therefore possible that I accidentally evolved rifampicin resistance in my donor. To check this future PCRs and sequencing of the rpoB gene could be performed to compare the sequence types of the mutations.
Methicillin resistance
I confirmed using a mecA PCR that the methicillin resistance gene was present in all of the colonies tested, and that all of my samples had high levels of rifampicin and oxacillin resistance. Overall, there are more experiments to do, to determine the frequency at which the SCCmec moves, but from my short placement it could be jumping between bacteria faster than we previously thought.
Having completed my placement, I have now gained the confidence to apply for PhD programmes and am actively searching for a PhD in microbiology here in the UK.
Find out more about AMI’s grants.





No comments yet