Artificial intelligence (AI) and “protein language” models can speed the design of monoclonal antibodies that prevent or reduce the severity of potentially life-threatening viral infections, according to a multi-institutional study led by researchers at Vanderbilt University Medical Center.
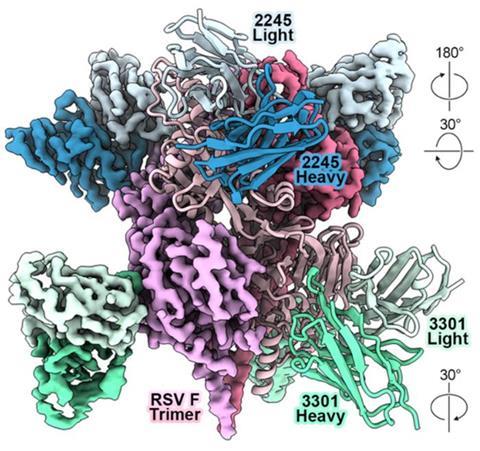
While their report, published Nov. 4 in the journal Cell, focused on development of antibody therapeutics against existing and emerging viral threats, including RSV (respiratory syncytial virus) and avian influenza viruses, the implications of the research are much broader, said the paper’s corresponding author, Ivelin Georgiev, PhD.
READ MORE: AI accelerates design of next-generation antimicrobial peptides with precision targeting
READ MORE: AI opens door to safe, effective new antibiotics to combat resistant bacteria
“This study is an important early milestone toward our ultimate goal — using computers to efficiently and effectively design novel biologics from scratch and translate them into the clinic,” said Georgiev, professor of Pathology, Microbiology and Immunology, and director of the Vanderbilt Program in Computational Microbiology and Immunology.
“Such approaches will have significant positive impact on public health and can be applied to a broad range of diseases, including cancer, autoimmunity, neurological diseases, and many others,” he said.
Georgiev is a leader in the use of computational approaches to advance disease treatment and prevention. He is the principal investigator of an up to $30 million award from the Advanced Research Projects Agency for Health (ARPA-H) to support the application of AI technology that can develop novel antibodies with therapeutic potential.
Perry Wasdin, PhD, a data scientist in the Georgiev lab, was involved in all aspects of the study and is first author of the paper.
Protein language model
The research team, which included scientists from around the country, Australia and Sweden, showed that a protein language model could design functional human antibodies that recognized the unique antigen sequencies (surface proteins) of specific viruses, without requiring part of the antibody sequence as a starting template.
Protein language models are a type of large language model (LLM), which is trained on huge amounts of text to enable language processing and generation. LLMs provide the core capabilities of chatbots such as ChatGPT.
By training their protein language model MAGE (Monoclonal Antibody Generator) on previously characterized antibodies against a known strain of the H5N1 influenza (bird flu) virus, the researchers were able to generate antibodies against a related, but unseen, influenza strain.
Emerging health threats
These findings suggest that MAGE “could be used to generate antibodies against an emerging health threat more rapidly than traditional antibody discovery methods,” which require blood samples from infected individuals or antigen protein from the novel virus, the researchers concluded.
Other Vanderbilt co-authors were Alexis Janke, PhD, Toma Marinov, PhD, Gwen Jordaan, Olivia Powers, Matthew Vukovich, PhD, Clinton Holt, PhD, and Alexandra Abu-Shmais.
This research was funded, in part, by the Advanced Research Projects Agency for Health (ARPA-H) and National Institutes of Health grants R01AI175245, R01AI152693, and 1ZIAAI005003. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the U.S. Government.
Topics
- Artificial Intelligence & Machine Learning
- Asia & Oceania
- avian influenza
- Computational Biology
- Emerging Threats & Epidemiology
- Immunology
- Infection Prevention & Control
- Infectious Disease
- Ivelin Georgiev
- One Health
- Perry Wasdin
- Pharmaceutical Microbiology
- protein language models
- Research News
- respiratory syncytial virus
- UK & Rest of Europe
- USA & Canada
- Vanderbilt University Medical Center
- Viruses



No comments yet